- assume_separate_talliesFalseWhether to assume that all tallies added in the XML files or by Cardinal are spatially separate. This is a performance optimization
Default:False
C++ Type:bool
Controllable:No
Description:Whether to assume that all tallies added in the XML files or by Cardinal are spatially separate. This is a performance optimization
- batch_interval1Trigger batch interval
Default:1
C++ Type:unsigned int
Controllable:No
Description:Trigger batch interval
- batchesNumber of batches to run in OpenMC; this overrides the setting in the XML files.
C++ Type:unsigned int
Controllable:No
Description:Number of batches to run in OpenMC; this overrides the setting in the XML files.
- calc_kinetics_paramsFalseWhether or not Cardinal should enable the calculation of kinetics parameters (Lambda effective and beta effective).
Default:False
C++ Type:bool
Controllable:No
Description:Whether or not Cardinal should enable the calculation of kinetics parameters (Lambda effective and beta effective).
- cell_levelCoordinate level in OpenMC (across the entire geometry) to use for identifying cells
C++ Type:unsigned int
Controllable:No
Description:Coordinate level in OpenMC (across the entire geometry) to use for identifying cells
- check_identical_cell_fillsFalseWhether to check that your model does indeed have identical cell fills, allowing you to set 'identical_cell_fills' to speed up initialization
Default:False
C++ Type:bool
Controllable:No
Description:Whether to check that your model does indeed have identical cell fills, allowing you to set 'identical_cell_fills' to speed up initialization
- check_tally_sumFalseWhether to check consistency between the local tallies with a global tally sum
Default:False
C++ Type:bool
Controllable:No
Description:Whether to check consistency between the local tallies with a global tally sum
- density_blocksBlocks corresponding to each of the 'density_variables'. If not specified, there will be no density feedback to OpenMC.
C++ Type:std::vector<std::vector<SubdomainName>>
Controllable:No
Description:Blocks corresponding to each of the 'density_variables'. If not specified, there will be no density feedback to OpenMC.
- density_variablesVector of variable names corresponding to the densities sent into OpenMC. Each entry maps to the corresponding entry in 'density_blocks.' If not specified, each entry defaults to 'density'
C++ Type:std::vector<std::vector<std::string>>
Controllable:No
Description:Vector of variable names corresponding to the densities sent into OpenMC. Each entry maps to the corresponding entry in 'density_blocks.' If not specified, each entry defaults to 'density'
- export_propertiesFalseWhether to export OpenMC's temperature and density properties to an HDF5 file after updating them from MOOSE.
Default:False
C++ Type:bool
Controllable:No
Description:Whether to export OpenMC's temperature and density properties to an HDF5 file after updating them from MOOSE.
- first_iteration_particlesNumber of particles to use for first iteration when using Dufek-Gudowski relaxation
C++ Type:int
Controllable:No
Description:Number of particles to use for first iteration when using Dufek-Gudowski relaxation
- identical_cell_fillsBlocks on which the OpenMC cells have identical fill universes; this is an optimization to speed up initialization for TRISO problems while also reducing memory usage. It is assumed that any cell which maps to one of these subdomains has exactly the same universe filling it as all other cells which map to these subdomains. We HIGHLY recommend that the first time you try using this, that you also set 'check_identical_cell_fills = true' to catch any possible user errors which would exclude you from using this option safely.
C++ Type:std::vector<SubdomainName>
Controllable:No
Description:Blocks on which the OpenMC cells have identical fill universes; this is an optimization to speed up initialization for TRISO problems while also reducing memory usage. It is assumed that any cell which maps to one of these subdomains has exactly the same universe filling it as all other cells which map to these subdomains. We HIGHLY recommend that the first time you try using this, that you also set 'check_identical_cell_fills = true' to catch any possible user errors which would exclude you from using this option safely.
- ifp_generations10The number of generations to use with the method of iterated fission probabilities.
Default:10
C++ Type:unsigned int
Controllable:No
Description:The number of generations to use with the method of iterated fission probabilities.
- inactive_batchesNumber of inactive batches to run in OpenMC; this overrides the setting in the XML files.
C++ Type:unsigned int
Controllable:No
Description:Number of inactive batches to run in OpenMC; this overrides the setting in the XML files.
- initial_propertiesmooseWhere to read the temperature and density initial conditions for OpenMC
Default:moose
C++ Type:MooseEnum
Controllable:No
Description:Where to read the temperature and density initial conditions for OpenMC
- k_triggernoneTrigger criterion to determine when OpenMC simulation is complete based on k
Default:none
C++ Type:MooseEnum
Controllable:No
Description:Trigger criterion to determine when OpenMC simulation is complete based on k
- k_trigger_thresholdThreshold for the k trigger
C++ Type:double
Unit:(no unit assumed)
Controllable:No
Description:Threshold for the k trigger
- lowest_cell_levelLowest coordinate level in OpenMC to use for identifying cells. The cell level for coupling will use the value set with this parameter unless the geometry does not have that many layers of geometry nesting, in which case the locally lowest depth is used
C++ Type:unsigned int
Controllable:No
Description:Lowest coordinate level in OpenMC to use for identifying cells. The cell level for coupling will use the value set with this parameter unless the geometry does not have that many layers of geometry nesting, in which case the locally lowest depth is used
- map_density_by_cellTrueWhether to apply a unique density to every OpenMC cell (the default), or instead apply a unique density to every OpenMC material (even if that material is filled into more than one cell). If your OpenMC model has a unique material in every cell you want to receive density feedback, these two options are IDENTICAL
Default:True
C++ Type:bool
Controllable:No
Description:Whether to apply a unique density to every OpenMC cell (the default), or instead apply a unique density to every OpenMC material (even if that material is filled into more than one cell). If your OpenMC model has a unique material in every cell you want to receive density feedback, these two options are IDENTICAL
- max_batchesMaximum number of batches, when using triggers
C++ Type:unsigned int
Controllable:No
Description:Maximum number of batches, when using triggers
- normalize_by_global_tallyTrueWhether to normalize local tallies by a global tally (true) or else by the sum of the local tally (false)
Default:True
C++ Type:bool
Controllable:No
Description:Whether to normalize local tallies by a global tally (true) or else by the sum of the local tally (false)
- openmc_verbosityOpenMC verbosity level; this overrides the setting in the XML files. Note that we cannot influence the verbosity of OpenMC's initialization routines, since these are run before Cardinal is initialized.
C++ Type:unsigned int
Controllable:No
Description:OpenMC verbosity level; this overrides the setting in the XML files. Note that we cannot influence the verbosity of OpenMC's initialization routines, since these are run before Cardinal is initialized.
- output_cell_mappingTrueWhether to automatically output the mapping from OpenMC cells to the [Mesh], usually for diagnostic purposes
Default:True
C++ Type:bool
Controllable:No
Description:Whether to automatically output the mapping from OpenMC cells to the [Mesh], usually for diagnostic purposes
- particlesNumber of particles to run in each OpenMC batch; this overrides the setting in the XML files.
C++ Type:unsigned int
Controllable:No
Description:Number of particles to run in each OpenMC batch; this overrides the setting in the XML files.
- powerPower (Watts) to normalize the OpenMC tallies; only used for k-eigenvalue mode
C++ Type:PostprocessorName
Unit:(no unit assumed)
Controllable:No
Description:Power (Watts) to normalize the OpenMC tallies; only used for k-eigenvalue mode
- regard_general_exceptions_as_errorsFalseIf we catch an exception during residual/Jacobian evaluaton for which we don't have specific handling, immediately error instead of allowing the time step to be cut
Default:False
C++ Type:bool
Controllable:No
Description:If we catch an exception during residual/Jacobian evaluaton for which we don't have specific handling, immediately error instead of allowing the time step to be cut
- relaxationnoneType of relaxation to apply to the OpenMC solution
Default:none
C++ Type:MooseEnum
Controllable:No
Description:Type of relaxation to apply to the OpenMC solution
- relaxation_factor0.5Relaxation factor for use with constant relaxation
Default:0.5
C++ Type:double
Unit:(no unit assumed)
Controllable:No
Description:Relaxation factor for use with constant relaxation
- reset_seedFalseWhether to reset OpenMC's seed to the initial starting seed before each OpenMC solve
Default:False
C++ Type:bool
Controllable:No
Description:Whether to reset OpenMC's seed to the initial starting seed before each OpenMC solve
- reuse_sourceFalseWhether to take the initial fission source for interation n to be the converged source bank from iteration n-1
Default:False
C++ Type:bool
Controllable:No
Description:Whether to take the initial fission source for interation n to be the converged source bank from iteration n-1
- scaling1Scaling factor to apply to [Mesh] to get to units of centimeters that OpenMC expects; setting 'scaling = 100.0', for instance, indicates that the [Mesh] is in units of meters
Default:1
C++ Type:double
Unit:(no unit assumed)
Controllable:No
Description:Scaling factor to apply to [Mesh] to get to units of centimeters that OpenMC expects; setting 'scaling = 100.0', for instance, indicates that the [Mesh] is in units of meters
- skinnerWhen using DAGMC geometries, an optional skinner that will regenerate the OpenMC geometry on-the-fly according to iso-contours of temperature and density
C++ Type:UserObjectName
Controllable:No
Description:When using DAGMC geometries, an optional skinner that will regenerate the OpenMC geometry on-the-fly according to iso-contours of temperature and density
- skip_statepointFalseWhether to skip writing any statepoint files from OpenMC; this is a performance optimization for scenarios where you may not want the statepoint files anyways
Default:False
C++ Type:bool
Controllable:No
Description:Whether to skip writing any statepoint files from OpenMC; this is a performance optimization for scenarios where you may not want the statepoint files anyways
- solveTrueWhether or not to actually solve the Nonlinear system. This is handy in the case that all you want to do is execute AuxKernels, Transfers, etc. without actually solving anything
Default:True
C++ Type:bool
Controllable:Yes
Description:Whether or not to actually solve the Nonlinear system. This is handy in the case that all you want to do is execute AuxKernels, Transfers, etc. without actually solving anything
- source_rate_normalizationScore to use for computing the particle source rate (source/sec) for a certain tallies in eigenvalue mode. In other words, the source/sec is computed as (power divided by the global value of this tally)
C++ Type:MooseEnum
Controllable:No
Description:Score to use for computing the particle source rate (source/sec) for a certain tallies in eigenvalue mode. In other words, the source/sec is computed as (power divided by the global value of this tally)
- source_strengthNeutrons/second to normalize the OpenMC tallies; only used for fixed source mode
C++ Type:PostprocessorName
Unit:(no unit assumed)
Controllable:No
Description:Neutrons/second to normalize the OpenMC tallies; only used for fixed source mode
- symmetry_mapperUser object (of type SymmetryPointGenerator) to map from a symmetric OpenMC model to a full-domain [Mesh]. For example, you can use this to map from a quarter-symmetric OpenMC model to a whole-domain [Mesh].
C++ Type:UserObjectName
Controllable:No
Description:User object (of type SymmetryPointGenerator) to map from a symmetric OpenMC model to a full-domain [Mesh]. For example, you can use this to map from a quarter-symmetric OpenMC model to a whole-domain [Mesh].
- tally_typeType of tally to use in OpenMC
C++ Type:MooseEnum
Controllable:No
Description:Type of tally to use in OpenMC
- temperature_blocksBlocks corresponding to each of the 'temperature_variables'. If not specified, there will be no temperature feedback to OpenMC.
C++ Type:std::vector<std::vector<SubdomainName>>
Controllable:No
Description:Blocks corresponding to each of the 'temperature_variables'. If not specified, there will be no temperature feedback to OpenMC.
- temperature_variablesVector of variable names corresponding to the temperatures sent into OpenMC. Each entry maps to the corresponding entry in 'temperature_blocks.' If not specified, each entry defaults to 'temp'
C++ Type:std::vector<std::vector<std::string>>
Controllable:No
Description:Vector of variable names corresponding to the temperatures sent into OpenMC. Each entry maps to the corresponding entry in 'temperature_blocks.' If not specified, each entry defaults to 'temp'
- verboseFalseWhether to print diagnostic information
Default:False
C++ Type:bool
Controllable:No
Description:Whether to print diagnostic information
- volume_calculationUser object that will perform a stochastic volume calculation to get the OpenMC cell volumes. This can be used to check that the MOOSE regions to which the cells map are of approximately the same volume as the true cells.
C++ Type:UserObjectName
Controllable:No
Description:User object that will perform a stochastic volume calculation to get the OpenMC cell volumes. This can be used to check that the MOOSE regions to which the cells map are of approximately the same volume as the true cells.
OpenMCCellAverageProblem
This class couples OpenMC cell-based models (e.g. Constructive Solid Geometry (CSG) or Direct Accelerated Geometry Monte Carlo (DAGMC)) to MOOSE. The crux is to identify a mapping between OpenMC cells and a MooseMesh. The data flow contains two major steps:
Temperature and/or density field data on the MooseMesh are volume-averaged and applied to the corresponding OpenMC cells.
Tallies are mapped from OpenMC into
CONSTANT MONOMIALfields on the MooseMesh through the tally system.
The smallest possible input file to run OpenMC is shown below. This page describes this syntax, plus more advanced settings.
Listing 1: Smallest possible OpenMC wrapped input file.
[Mesh<<<{"href": "../../syntax/Mesh/index.html"}>>>]
type = FileMesh
file = pincell.e
[]
[Problem<<<{"href": "../../syntax/Problem/index.html"}>>>]
type = OpenMCCellAverageProblem
power = 1000.0
temperature_blocks = '1 2 3'
density_blocks = '3'
cell_level = 0
[Tallies<<<{"href": "../../syntax/Problem/Tallies/index.html"}>>>]
[Cell]
type = CellTally<<<{"description": "A class which implements distributed cell tallies.", "href": "../tallies/CellTally.html"}>>>
blocks<<<{"description": "This parameter is deprecated, use 'block' instead!"}>>> = '1'
[]
[]
[]
[Executioner<<<{"href": "../../syntax/Executioner/index.html"}>>>]
type = Transient
[]
[Outputs<<<{"href": "../../syntax/Outputs/index.html"}>>>]
exodus<<<{"description": "Output the results using the default settings for Exodus output."}>>> = true
[]Initializing Variables
We first initialize (MooseVariables) to communicate OpenMC's solution with MOOSE. These variables will be viewable in the MOOSE output files (e.g., via Paraview). Depending on the user settings, the following CONSTANT MONOMIAL variables will be added:
Variable(s) representing the OpenMC tally(s) added by the
[Tallies]blockVariable(s) representing the temperature to read into OpenMC. Temperature will be read from the mesh subdomains indicated by
temperature_blocks.Variable(s) representing the density to read into OpenMC. Density will be read from the mesh subdomains indicated by
density_blocks.A variable (
cell_id) storing the mapping of OpenMC cell IDs onto the meshA variable (
cell_instance) storing the mapping of OpenMC cell instances onto the mesh
These variables have default names, but you can also control their names using the temperature_variables, and density_variables parameters. The names of the tally variables can be customized with the tally_name parameter in each tally object. If you want to see the names, run with verbose = true and tables will print out on initialization with this information.
You do not need to add/initialize ANY of these variables manually - it all happens behind the scenes!
As an example, let's start with a complicated case first. Suppose our [Problem] block looks like the following. We want to apply temperature feedback from four blocks (fuel, cladding, water, helium). We also want to apply density feedback, but only from two blocks (water and fluid). Suppose we also want to keep separate track of different variables representing each of these physical fields, so we will want to read temperature from a variable named temp0 in the fuel and cladding blocks, but from a variable named nek_temp in the helium block (and so on for density).
[Problem]
type = OpenMCCellAverageProblem
temperature_variables = 'temp0; t_water; nek_temp'
temperature_blocks = 'fuel cladding; water; helium'
density_variables = 'rho_water; rho_helium'
density_blocks = 'water; helium'
[Tallies]
[cell_tally]
type = CellTally
score = 'heating flux'
name = 'power openmc_flux'
[]
[]
[]
Then Cardinal is building the following automatically for you:
[AuxVariables]
[temp0] # added for all corresponding 'temperature_blocks'
family = MONOMIAL
order = CONSTANT
block = 'fuel cladding'
[]
[t_water] # added for all corresponding 'temperature_blocks'
family = MONOMIAL
order = CONSTANT
block = 'water'
[]
[nek_temp] # added for all corresponding 'temperature_blocks'
family = MONOMIAL
order = CONSTANT
block = 'helium'
[]
[rho_water] # added for all corresponding 'density_blocks'
family = MONOMIAL
order = CONSTANT
blocks = 'water'
[]
[rho_helium] # added for all corresponding 'density_blocks'
family = MONOMIAL
order = CONSTANT
blocks = 'helium'
[]
[power] # the first tally we added (score is 'heating', but we set a custom name)
family = MONOMIAL
order = CONSTANT
[]
[openmc_flux] # the second tally we added (score is 'flux', but we set a custom name)
family = MONOMIAL
order = CONSTANT
[]
[]
By default, Cardinal will name all temperature feedback with the temp variable, all density feedback with the density variable, and all tally scores with the same name as the score. Suppose we instead wanted to rely on defaults; we would set our [Problem] block as:
[Problem]
type = OpenMCCellAverageProblem
density_blocks = 'water helium'
temperature_blocks = 'fuel cladding water helium'
[Tallies]
[cell_tally]
type = CellTally
score = 'heating flux'
[]
[]
[]
Then Cardinal is instead building the following automatically for you:
[AuxVariables]
[temp] # added for all temperature_blocks
family = MONOMIAL
order = CONSTANT
block = 'fuel cladding water helium'
[]
[density] # added for all density_blocks
family = MONOMIAL
order = CONSTANT
block = 'water helium'
[]
[heating] # the first tally we added (score is 'heating')
family = MONOMIAL
order = CONSTANT
[]
[flux] # the second tally we added (score is 'flux')
family = MONOMIAL
order = CONSTANT
[]
[]
Cell to Element Mapping
Next, a mapping from the elements in the MooseMesh (i.e. the mesh in the [Mesh] block) is established to the OpenMC cell geometry.
The OpenMC geometry is always specified in length units of centimeters. Most other MOOSE applications use SI units, i.e. meters for the length unit. See Mesh Scaling to learn how to couple OpenMC models to non-centimeter-based MOOSE applications.
In the [Mesh] block, you should provide a mesh onto which OpenMC will write its tally values, as well as read temperature and density. This mesh is only used for reading/writing data, so there are no requirements on node continuity across elements. We loop over all the elements and map each to an OpenMC cell according to the element centroid. As an example, Figure 1 depicts an OpenMC geometry, a mesh on which temperature feedback is to be applied, and a visualization of the mapping from these elements to the OpenMC cells. There are no requirements on alignment of elements/cells or on preserving volumes - the OpenMC cells and mesh mirror elements do not need to be conformal. Elements that don't map to an OpenMC cell simply do not participate in the multiphysics coupling (and vice versa for the cells). This feature can be used to exclude regions such as reflectors from multiphysics feedback.
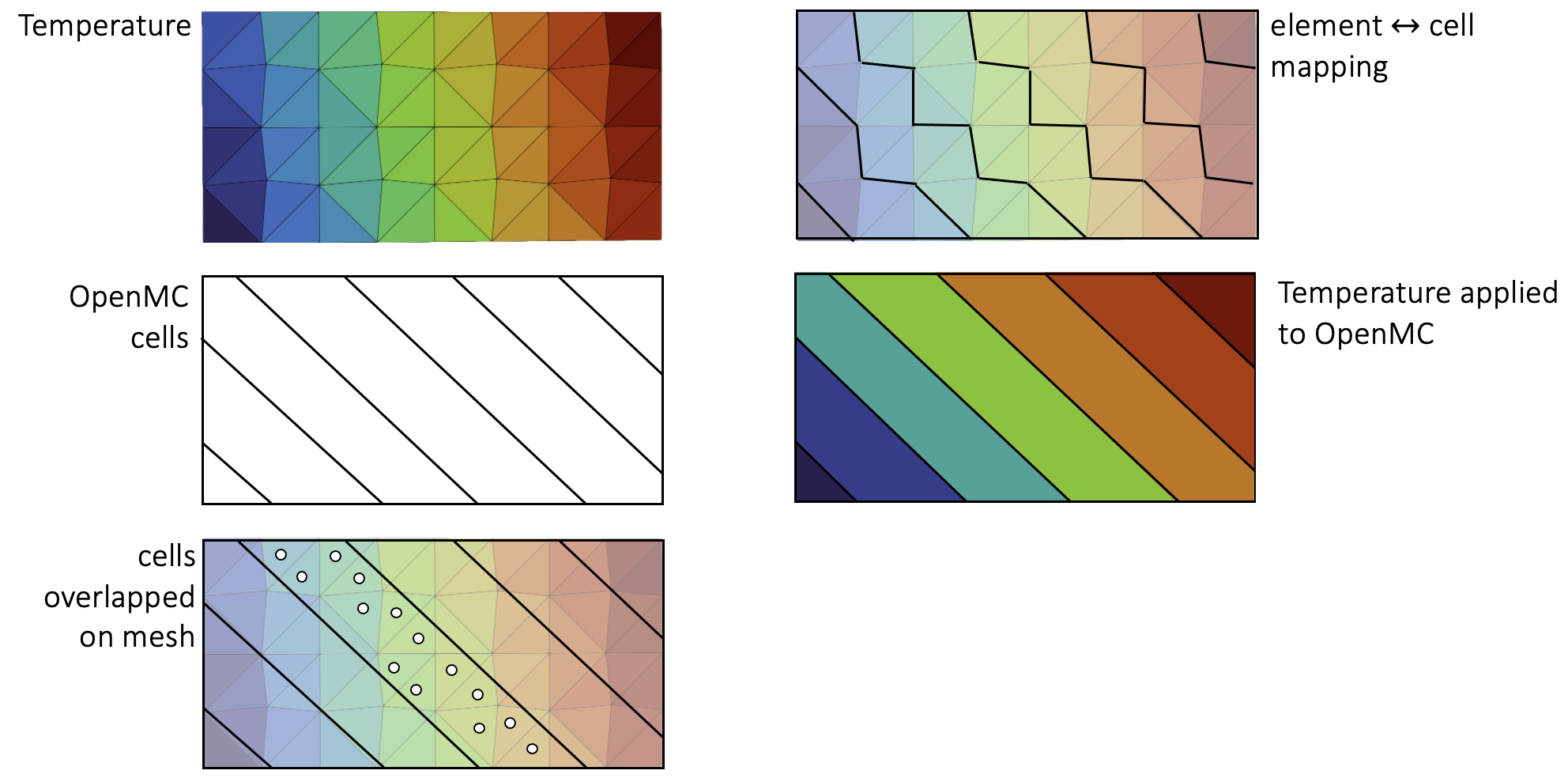
Figure 1: Illustration of OpenMC cells and the mapping to a mesh.
You can visualize how the OpenMC cells map to the mesh by viewing the cell_id and cell_instance variables Cardinal automatically outputs. For more information on how to interpret these variables, see: CellIDAux and CellInstanceAux.
The cell-to-element mapping should be established with care. There are two general behavior patterns that are typically undesirable. In Figure 2, consider the case where the [Mesh] has four equal-sized elements, each of volume , while the OpenMC domain has three equal-sized cells. Assume that the power produced by each OpenMC cell is , so that the total power of the OpenMC domain is . By nature of the centroid mapping, one OpenMC cell will map to a much larger region of space than the other two cells. Even though the fission power in each OpenMC cell is , the power density will differ by a factor of two once mapped to the [Mesh]. In practice, this might not have a detrimental or even noticeable effect on temperature solutions due to the smoothing nature of heat conduction, but you should be aware of it.
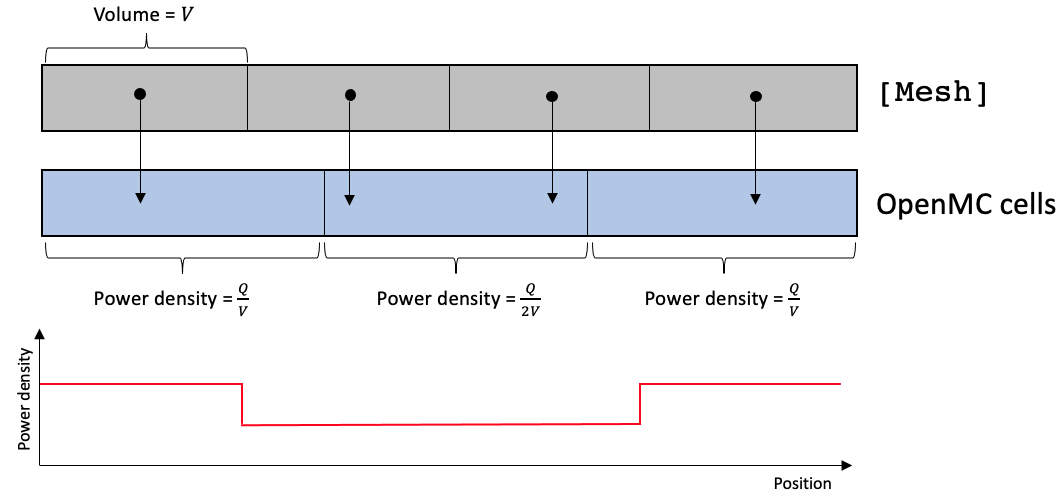
Figure 2: Element to cell mapping for OpenMC cells coarser than the [Mesh]
You can monitor the mapping by checking the mesh volumes that each OpenMC cell maps to by setting verbose = true, which will print a mapping table. You can also add an OpenMCVolumeCalculation which will run a stochastic volume calculation and print out the actual volume of the OpenMC cells in the "Actual Vol" column (not just the volume of the mesh elements the cells map to in the "Mapped Vol" column!).
---------------------------------------------------------------------------
| Cell | T | T+rho | Other | Mapped Vol | Actual Vol |
---------------------------------------------------------------------------
| 1, instance 0 (of 10) | 168 | 0 | 0 | 5.298e-01 | |
| 1, instance 1 (of 10) | 168 | 0 | 0 | 5.298e-01 | |
| 1, instance 2 (of 10) | 168 | 0 | 0 | 5.298e-01 | |
---------------------------------------------------------------------------
If you know that all the tallied OpenMC cells are actually the same volume, you can also use the check_equal_mapped_tally_volumes parameter to automatically check that the mapped volue for each tally bin is identical.
In Figure 3, consider the case where the [Mesh] has two equal-sized elements, while the OpenMC domain has three equal-sized cells. By nature of the centroid mapping, one OpenMC cell will not participate in coupling because no elements in the [Mesh] have a centroid within the cell. This situation is more difficult to detect, but can be monitored by setting verbose = true and monitoring the table outputs.
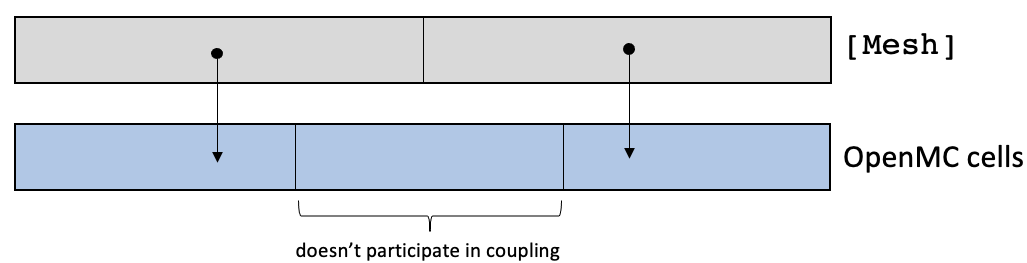
Figure 3: Element to cell mapping for OpenMC cells finer than the [Mesh]
Of course, the situations shown in Figure 2 and Figure 3 can occur even if the number of mesh elements exactly match the number of OpenMC cells, depending on where the element centroids are located. Therefore, we recommend using verbose = true when getting started.
Cell Levels
A unique temperature and density will be assigned to each OpenMC cell that has a unique combination of a cell ID and instance. As such, the cell "level" on which to apply multiphysics feedback must be defined, using either the cell_level or lowest_cell_level parameter.
The cell "level" is the coordinate "level" in the OpenMC geometry to "stop at" for identifying the cell ID/instance pairs. Cell-based geometries can be constructed by nesting repeated universes/lattices at multiple locations in the domain, but you may not be interested in always coupling cells at the lowest level to MOOSE. This feature was added to Cardinal specifically for TRISO applications, where heat conduction feedback is usually not explicitly resolving TRISO particles (instead, you'd prefer to set each fuel pebble or compact to a single temperature, requiring Cardinal to figure out the list of cells (TRISO layers, matrix) filling some higher-level cell (the pebble surface, the compact cylinder surface). The cell_level and lowest_cell_level parameters indicate the coordinate level (relative to 0, the highest level in the geometry) at which to "stop" the find cell routine.
You can visualize your OpenMC model at different geometry hierarchies using OpenMC plots and setting the level.
As an example, the figure below shows a TRISO-fueled High Temperature Gas Reactor (HTGR) fuel assembly from the gas assembly tutorial. This image shows the geomtery, colored by cell, at different levels in the geometry. If we wanted to apply a single temperature value to all the TRISO particles nested inside a compact, we would set cell_level = 1 for this problem. If we chose cell_level = 2, we'd be trying to set a unique temperature for every individual TRISO particle. If we chose cell_level = 3, we'd be trying to set a unique temperature for every layer in every TRISO particle.
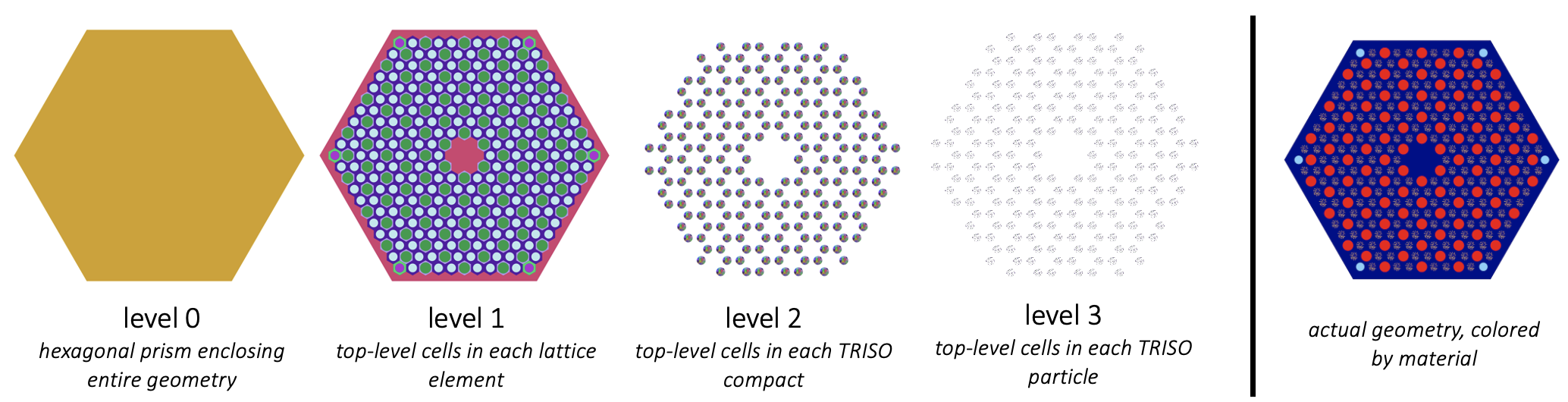
Figure 4: Cell level concept, using a TRISO assembly as an example
In many cases, the coordinate levels on which you would like to couple OpenMC to MOOSE are not the same everywhere in the OpenMC geometry. This was the case in Figure 4 (levels 2 and 3 only exist in parts of the domain). You can use lowest_cell_level to indicate that coupling should be performed on the specified level except in regions where there are no cells at that level.
Adding Tallies
Mapped Tallies
This class takes the tally objects initialized by the [Tallies] block and uses them to construct tally auxvariables. At the moment there are two options for discretizing tallies spatially in Cardinal:
If no tallies are specified by the [Tallies] block, this class adds no tally auxvariables. Each is normalized according to the specified power or source_strength (depending on whether you are running a -eigenvalue or fixed-source problem). By default, the normalization is done against a global tally added over the entire OpenMC domain. By setting normalize_by_global_tally to false, however, the tally is instead normalized by the sum of the tally itself.
Mapped Tally Scores
You can customize the type of score that Cardinal uses to normalize tallies to power with the source_rate_normalization parameter. Options include:
heating: total nuclear heatingheating_local: same as theheatingscore, except that energy from secondary photons is deposited locallykappa_fission: recoverable energy from fission, including prompt sources (fission fragments, prompt neutrons, prompt gammas) and delayed sources (delayed neutrons, delayed gammas, delayed betas). Neutrino energy is neglected. The energy from photons is assumed to deposit locally.fission_q_prompt: the prompt components of thekappa_fissionscore, except that the energy released is a function of the incident energy by linking to optional fission energy release data.fission_q_recoverable: same as thekappa_fissionscore, except that the score depends on the incident energy by linking to optional fission energy release data
For more information on the specific meanings of these various scores, please consult the OpenMC tally documentation. All of the tallies added are normalized with the same source_rate_normalization score when running in eigenvalue mode. Table 1 compares the units from OpenMC and the units of the AuxVariables created for all tally scores supported by Cardinal. Note that for all area or volume units in Table 1, that those units match whatever unit is used in the [Mesh].
Table 1: Tally units from OpenMC and the conversion in Cardinal.
| Tally score | OpenMC Units | Cardinal Units |
|---|---|---|
heating | eV / source particle | W / volume |
heating_local | eV / source particle | W / volume |
kappa_fission | eV / source particle | W / volume |
fission_q_prompt | eV / source particle | W / volume |
fission_q_recoverable | eV / source particle | W / volume |
damage_energy | eV / source particle | eV / volume / second |
flux | particle - cm / source particle | particle / area / second |
H3_production | tritium / source particle | tritium / volume / second |
total | total reactions / source particle | total reactions / volume / second |
absorption | absorption reactions / source particle | absorption reactions / volume / second |
scatter | scattering reactions / source particle | scattering reactions / volume / second |
nu_scatter | nu-scattering reactions / source particle | nu-scattering reactions / volume / second |
fission | fission reactions / source particle | fission reactions / volume / second |
nu_fission | nu-fission reactions / source particle | nu-fission reactions / volume / second |
inverse_velocity | seconds / source particle | particle / volume |
Tally Normalization
The tally units-transformation process involves division by a volume. In Cardinal, there are two different notions of volume:
The volume of the
[Mesh]elements which map to a tally bin regionThe actual volume of the tally bin region in OpenMC
Take Figure 5 as an example; on the left is a [Mesh], and on the right is a spherical OpenMC cell we are mapping to the [Mesh]. For this case, the two volumes are different, because of the faceted nature of the sphere surface. The tallies in Cardinal are all normalized by the [Mesh] volume (0.96 cm in this example). This is a good choice for heating-related tallies, because this will ensure we properly preserve a total integral (power) when integrated on the [Mesh]. For heating tallies, it is much more important to preserve the total power, as opposed to the raw pointwise value of the heat deposition (units of W/m).
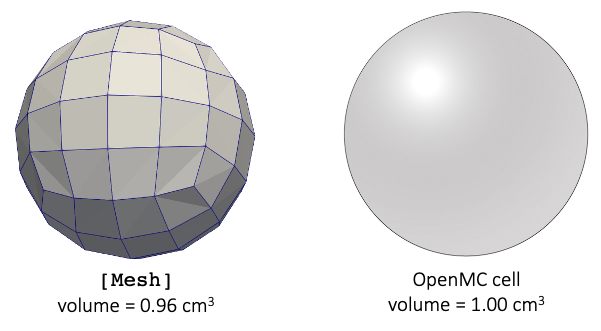
Figure 5: Illustration of OpenMC particle transport geometry and the mapping of OpenMC cells to a user-supplied "mesh mirror."
However, for some other tallies (e.g. the 'flux' tallies), dividing by the [Mesh] volume indicates instead that we're normalizing so as to preserve a reaction rate (reactions/sec), as if you integrated over the [Mesh]. This will not give the same raw pointwise tally value as what OpenMC is actually predicting, if the volumes of the [Mesh] and the tally bin regions are significantly different. If the pointwise value is very important for your use case, you can post-multiply your AuxVariables by (in this example, 0.96 / 1.00), where is the volume of the [Mesh] elements which map to the tally bin, and is the volume of the corresponding tally region.
If your OpenMC tally bins and corresponding [Mesh] elements already are exactly the same volume, then no special thought is needed for the tally normalization, and the value will be exactly consistent with the interpretation used in OpenMC.
Tally and Filter Editors
Cardinal provides UserObjects for editing tallies and filters:
- OpenMCTallyEditor: online control of tally parameters - OpenMCDomainFilterEditor: online control of domain filter parameters
These objects can be used to interact with tallies and/or filters that are present in a tallies.xml file for the problem being run (in other words, tallies the user manually sets up when building the OpenMC model). These objects can be used to create tallies and/or filters as well. In either case, these objects do not interact with tallies and filters used to map data to the mesh mirror (Mapped Tallies ).
Neutron Kinetics Parameters
Cardinal exposes the calculation of neutron kinetics parameters in OpenMC with iterated fission probabilities. and can be obtained by first setting calc_kinetics_params = true. The parameters can then be accessed with LambdaEffective and BetaEffective post-processors after the OpenMC simulation finishes. An example of this capability can be found below.
[Problem<<<{"href": "../../syntax/Problem/index.html"}>>>]
type = OpenMCCellAverageProblem
verbose = true
calc_kinetics_params = true
ifp_generations = 5
[][Postprocessors<<<{"href": "../../syntax/Postprocessors/index.html"}>>>]
[lambda]
type = LambdaEffective<<<{"description": "A post-processor which computes and returns the kinetics parameter $\\Lambda_{eff}$.", "href": "../postprocessors/LambdaEffective.html"}>>>
[]
[lambda_rel]
type = LambdaEffective<<<{"description": "A post-processor which computes and returns the kinetics parameter $\\Lambda_{eff}$.", "href": "../postprocessors/LambdaEffective.html"}>>>
output<<<{"description": "The value to output. Options are $\\Lambda_{eff}$ (mean), the standard deviation of $\\Lambda_{eff}$ (std_dev), or the relative error of $\\Lambda_{eff}$ (rel_err)."}>>> = rel_err
[]
[lambda_std]
type = LambdaEffective<<<{"description": "A post-processor which computes and returns the kinetics parameter $\\Lambda_{eff}$.", "href": "../postprocessors/LambdaEffective.html"}>>>
output<<<{"description": "The value to output. Options are $\\Lambda_{eff}$ (mean), the standard deviation of $\\Lambda_{eff}$ (std_dev), or the relative error of $\\Lambda_{eff}$ (rel_err)."}>>> = std_dev
[]
[beta]
type = BetaEffective<<<{"description": "A post-processor which computes and returns the kinetics parameter $\\beta_{eff}$.", "href": "../postprocessors/BetaEffective.html"}>>>
[]
[beta_rel]
type = BetaEffective<<<{"description": "A post-processor which computes and returns the kinetics parameter $\\beta_{eff}$.", "href": "../postprocessors/BetaEffective.html"}>>>
output<<<{"description": "The value to output. Options are $\\beta_{eff}$ (mean), the standard deviation of $\\beta_{eff}$ (std_dev), or the relative error of $\\beta_{eff}$ (rel_err)."}>>> = rel_err
[]
[beta_std]
type = BetaEffective<<<{"description": "A post-processor which computes and returns the kinetics parameter $\\beta_{eff}$.", "href": "../postprocessors/BetaEffective.html"}>>>
output<<<{"description": "The value to output. Options are $\\beta_{eff}$ (mean), the standard deviation of $\\beta_{eff}$ (std_dev), or the relative error of $\\beta_{eff}$ (rel_err)."}>>> = std_dev
[]
[]The number of generations tracked to compute these kinetic parameters can be specified by setting ifp_generations. Note that ifp_generations must be less than or equal to the number of inactive batches.
The memory cost of a simulation running with iterated fission probabilities scales linearly with ifp_generations (an extra double and an extra integrer must be tracked per particle for each generation). It is recommend that the number of IFP generations be minimized when possible.
Other Features
While this class mainly facilitates data transfers to and from OpenMC, a number of other features are implemented.
CAD Geometry Skinning
OpenMC is currently limited to setting a single constant temperature and a single constant density in each cell. For CSG geometries, the user needs to manually set up sub-divisions in the geometry in order to capture spatial variation in temperature and density. For DAGMC geometries, you can instead optionally re-generate the OpenMC cells after each Picard iteration according to contours in temperature and/or density. With this approach, the OpenMC model can receive spatially-varying temperature and density without the user needing to manually subdivide regions of space a priori. To use this feature, set the skinner parameter to the name of a MoabSkinner user object. For more information, consult the documentation for MoabSkinner.
This skinning feature is only available for DAGMC geometries for which the [Mesh] obeys the same material boundaries in the starting .h5m file.
Mesh Scaling
OpenMC always uses a length unit of centimeters, but a coupled MOOSE application often uses a length unit of meters. When transferring field data, it is important to account for the length units of the coupled MOOSE application. The scaling parameter is used to apply a multiplicative factor to the [Mesh] and mesh_template (if using a file-based mesh tally) to get to units of centimeters assumed by OpenMC. This multiplicative factor is then used to
"Find cell" routines in OpenMC in order to map a centimeters-based OpenMC geometry to a different length unit`
Scale the fission power in OpenMC (units of W/cm) to a different volume unit
If an unstructured mesh tally is used, scale the
mesh_templateandmesh_translations/mesh_translations_fileto units of centimeters
For instance, if the [Mesh] is specified in units of meters, then scaling should be set to 100.0 to indicate that the [Mesh] is specified in a unit 100.0 times larger than the OpenMC unit of centimeters.
Tally Optimizations
"Spatially separate" tallies are tallies where a particle can only score to one bin for a given event - for instance, a tally with one bin per fuel pebble in a pebble bed reactor is a spatially separate tally because a particle scoring to fission in pebble cannot also score to fission in pebble . OpenMC allows you to specify that tallies are spatially separate, which can offer a big performance improvement. If your OpenMC input files do not contain any of their own tallies (i.e. in a tallies.xml), you can try using this feature to speed up your calculation.
Relaxation
OpenMC is coupled to MOOSE via fixed point iteration, also referred to as Picard iteration. For many problems, instabilities can exist between OpenMC and the coupled thermal-fluid physics. The OpenMC wrapping offers several options by which the tally can be "relaxed" between iterations to effectively damp these oscillations. For all these relaxation schemes, the tally that gets coupled to MOOSE for iteration is taken as a weighted sum of the previous iterate and the most-recent Monte Carlo calculation:
(1)
where is the weighting factor, indicates the -th Monte Carlo solve, and the number of samples is . For the very first fixed point iteration, because there is no previous iterate, . For , Eq. (1) is referred to as "over-relaxation" because more than a full step is taken in the direction of the most recent iterate. Conversely, for , Eq. (1) is referred to as "under-relaxation" because less than a full step is taken in the direction of the most recent iterate. Several options are available for selecting and . These are chosen with the relaxation parameter. Options include:
constant: choose a constant androbbins_monro: and constant ; applying the recursion in Eq. (1) shows that the solution at iteration is the average of the previous Monte Carlo solutions
(2)
dufek_gudowski: variable and ; the step size is selected based on the fraction of samples in the -th Monte Carlo solve relative to the total number of samples performed so far. The number of samples is then based on the notion that the step size should be proportional to the convergence rate of the central limit theorem (inversely proportional to the square root of the total number of simulated particles):
(3)
Controlling OpenMC Termination
OpenMC's tally triggers allow OpenMC to automatically end its active batches once reaching certain criteria in and/or the tally uncertainties, including:
standard deviation
variance
relative error
tally relative error
Setting k_trigger enables triggers based on , and setting k_trigger_threshold sets the required convergence criteria. Individual tally triggers can be set in the tally_trigger and tally_trigger_threshold parameters of the tally classes (CellTally and MeshTally). Both and multiple tally triggers can be used simultaneously.
Controlling the OpenMC Settings
This class provides minimal capabilities to control the OpenMC simulation settings directly from the Cardinal input file:
batches: the number of batches (inactive plus active)inactive_batches: the number of inactive batchesopenmc_verbosity: verbosity level in OpenMCparticles: the number of particles per batch
For all of the above, a setting in the Cardinal input file will override any settings in the OpenMC XML files.
Volume Calculations
It can be helpful for debugging problem setup to compare actual OpenMC cell volumes against the [Mesh] element volumes to which they map. A well-designed mapping should have good agreement between actual cell volumes and the [Mesh] volumes they map to. To add a volume calculation, set the volume_calculation parameter to the name of a OpenMCVolumeCalculation object. If you then set verbose = true, you will be able to compare the cell volumes with the MOOSE elements.
Symmetric Data Transfers
Many neutronics problems are symmetric - such as half-core or quarter-core symmetry. However, some MOOSE mesh generators don't yet support symmetric geometries by slicing along arbitrary planes. This class supports both half-symmetric OpenMC models and general -th symmetric rotational models by coupling symmetric OpenMC models to full-system thermal-fluid models.
The symmetric mapping is specified with the symmetry_mapper parameter, a SymmetryPointGenerator user object. Note that if asymmetries exist in whatever physics OpenMC is coupled to, they will be averaged on both sides of the plane before sending temperatures and densities to OpenMC. Please consult the SymmetryPointGenerator for more information.
Input Parameters
- allow_initial_conditions_with_restartFalseTrue to allow the user to specify initial conditions when restarting. Initial conditions can override any restarted field
Default:False
C++ Type:bool
Controllable:No
Description:True to allow the user to specify initial conditions when restarting. Initial conditions can override any restarted field
- restart_file_baseFile base name used for restart (e.g.
/ or /LATEST to grab the latest file available) C++ Type:FileNameNoExtension
Controllable:No
Description:File base name used for restart (e.g.
/ or /LATEST to grab the latest file available)
Restart Parameters
- control_tagsAdds user-defined labels for accessing object parameters via control logic.
C++ Type:std::vector<std::string>
Controllable:No
Description:Adds user-defined labels for accessing object parameters via control logic.
- default_ghostingFalseWhether or not to use libMesh's default amount of algebraic and geometric ghosting
Default:False
C++ Type:bool
Controllable:No
Description:Whether or not to use libMesh's default amount of algebraic and geometric ghosting
- enableTrueSet the enabled status of the MooseObject.
Default:True
C++ Type:bool
Controllable:No
Description:Set the enabled status of the MooseObject.
Advanced Parameters
- kernel_coverage_block_listList of subdomains for kernel coverage check. The meaning of this list is controlled by the parameter 'kernel_coverage_check' (whether this is the list of subdomains to be checked, not to be checked or not taken into account).
C++ Type:std::vector<SubdomainName>
Controllable:No
Description:List of subdomains for kernel coverage check. The meaning of this list is controlled by the parameter 'kernel_coverage_check' (whether this is the list of subdomains to be checked, not to be checked or not taken into account).
- material_coverage_block_listList of subdomains for material coverage check. The meaning of this list is controlled by the parameter 'material_coverage_check' (whether this is the list of subdomains to be checked, not to be checked or not taken into account).
C++ Type:std::vector<SubdomainName>
Controllable:No
Description:List of subdomains for material coverage check. The meaning of this list is controlled by the parameter 'material_coverage_check' (whether this is the list of subdomains to be checked, not to be checked or not taken into account).
- material_coverage_checkTRUEControls, if and how a material subdomain coverage check is performed. With 'TRUE' or 'ON' all subdomains are checked (the default). Setting 'FALSE' or 'OFF' will disable the check for all subdomains. To exclude a predefined set of subdomains 'SKIP_LIST' is to be used, while the subdomains to skip are to be defined in the parameter 'material_coverage_block_list'. To limit the check to a list of subdomains, 'ONLY_LIST' is to be used (again, using the parameter 'material_coverage_block_list').
Default:TRUE
C++ Type:MooseEnum
Controllable:No
Description:Controls, if and how a material subdomain coverage check is performed. With 'TRUE' or 'ON' all subdomains are checked (the default). Setting 'FALSE' or 'OFF' will disable the check for all subdomains. To exclude a predefined set of subdomains 'SKIP_LIST' is to be used, while the subdomains to skip are to be defined in the parameter 'material_coverage_block_list'. To limit the check to a list of subdomains, 'ONLY_LIST' is to be used (again, using the parameter 'material_coverage_block_list').
Simulation Checks Parameters
- not_zeroed_tag_vectorsExtra vector tags which the sytem will not zero when other vector tags are zeroed. The outer index is for which nonlinear system the extra tag vectors should be added for
C++ Type:std::vector<std::vector<TagName>>
Controllable:No
Description:Extra vector tags which the sytem will not zero when other vector tags are zeroed. The outer index is for which nonlinear system the extra tag vectors should be added for
Contribution To Tagged Field Data Parameters
- parallel_barrier_messagingFalseDisplays messaging from parallel barrier notifications when executing or transferring to/from Multiapps (default: false)
Default:False
C++ Type:bool
Controllable:No
Description:Displays messaging from parallel barrier notifications when executing or transferring to/from Multiapps (default: false)
- verbose_multiappsFalseSet to True to enable verbose screen printing related to MultiApps
Default:False
C++ Type:bool
Controllable:No
Description:Set to True to enable verbose screen printing related to MultiApps
- verbose_restoreFalseSet to True to enable verbose screen printing related to solution restoration
Default:False
C++ Type:bool
Controllable:No
Description:Set to True to enable verbose screen printing related to solution restoration
- verbose_setupfalseSet to 'true' to have the problem report on any object created. Set to 'extra' to also display all parameters.
Default:false
C++ Type:MooseEnum
Controllable:No
Description:Set to 'true' to have the problem report on any object created. Set to 'extra' to also display all parameters.
Verbosity Parameters
- restore_original_nonzero_patternFalseWhether we should reset matrix memory for every Jacobian evaluation. This option is useful if the sparsity pattern is constantly changing and you are using hash table assembly or if you wish to continually restore the matrix to the originally preallocated sparsity pattern computed by relationship managers.
Default:False
C++ Type:bool
Controllable:No
Description:Whether we should reset matrix memory for every Jacobian evaluation. This option is useful if the sparsity pattern is constantly changing and you are using hash table assembly or if you wish to continually restore the matrix to the originally preallocated sparsity pattern computed by relationship managers.
- use_hash_table_matrix_assemblyFalseWhether to assemble matrices using hash tables instead of preallocating matrix memory. This can be a good option if the sparsity pattern changes throughout the course of the simulation.
Default:False
C++ Type:bool
Controllable:No
Description:Whether to assemble matrices using hash tables instead of preallocating matrix memory. This can be a good option if the sparsity pattern changes throughout the course of the simulation.
Nonlinear System(S) Parameters
- show_invalid_solution_consoleTrueSet to true to show the invalid solution occurance summary in console
Default:True
C++ Type:bool
Controllable:No
Description:Set to true to show the invalid solution occurance summary in console
Solution Validity Control Parameters
Input Files
- (test/tests/neutronics/tallies/tritium/openmc.i)
- (test/tests/neutronics/filters/azimuthal/cell.i)
- (test/tests/openmc_errors/no_range/openmc.i)
- (test/tests/openmc_errors/incorrect_var_setup/incorrect_var_type.i)
- (test/tests/neutronics/symmetry/rotational/openmc.i)
- (test/tests/neutronics/feedback/universes/lattice_in_flat/openmc.i)
- (test/tests/neutronics/mesh_tally/fission_tally_std_dev.i)
- (test/tests/openmc_errors/block_mappings/skipping_moose_feedback.i)
- (test/tests/neutronics/heat_source/custom_name.i)
- (test/tests/symmetry/reflection.i)
- (test/tests/openmc_errors/block_mappings/multiple_phases.i)
- (test/tests/openmc_errors/mesh_tally/incorrect_order.i)
- (test/tests/neutronics/tallies/reaction_rates/openmc.i)
- (test/tests/neutronics/fixed_source/overlap_all.i)
- (test/tests/neutronics/tally_system/multi_diff.i)
- (test/tests/neutronics/dagmc/with_csg/multi_cell_dag/openmc.i)
- (test/tests/neutronics/filters/polar/mesh.i)
- (test/tests/openmc_errors/fixed_source/k_trigger.i)
- (test/tests/openmc_errors/block_mappings/unequal_volumes.i)
- (test/tests/neutronics/dagmc/mesh_tallies/one_bin.i)
- (test/tests/neutronics/feedback/universes/fill_in_flat/openmc.i)
- (test/tests/neutronics/feedback/multi_component_density/multi/openmc.i)
- (test/tests/neutronics/heat_source/multi_tally_mesh.i)
- (tutorials/lwr_mgxs/openmc_mgxs.i)
- (test/tests/neutronics/triggers/ignore_zeros/ignore_zero.i)
- (test/tests/neutronics/openmc_xml/openmc.i)
- (test/tests/neutronics/feedback/interchangeable/no_data_flow.i)
- (test/tests/neutronics/dagmc/mesh_tallies/direct_tally.i)
- (test/tests/openmc_errors/block_mappings/no_overlap.i)
- (test/tests/userobjects/volume_calculation/instances/openmc.i)
- (test/tests/neutronics/feedback/multi_component_temp/openmc_incorrect_length.i)
- (test/tests/neutronics/symmetry/wrong_uo.i)
- (test/tests/neutronics/feedback/single_level/openmc.i)
- (test/tests/neutronics/dagmc/mesh_tallies/openmc.i)
- (test/tests/neutronics/mesh_tally/multiple_meshes.i)
- (test/tests/neutronics/kinetics/non_eigen/openmc.i)
- (test/tests/neutronics/dagmc/with_csg/dag_cell_not_root/openmc.i)
- (test/tests/neutronics/indicators/od/openmc.i)
- (test/tests/neutronics/gen_mgxs/all_mgxs_cell_l1.i)
- (test/tests/userobjects/openmc_nuclide_densities/no_change.i)
- (test/tests/neutronics/tallies/reaction_rates/openmc_mgxs_rxn.i)
- (test/tests/neutronics/fixed_source/overlap_solid.i)
- (test/tests/neutronics/tally_system/cell_with_unmapped.i)
- (test/tests/neutronics/filters/legendre/cell.i)
- (test/tests/neutronics/filters/particle/cell.i)
- (test/tests/userobjects/volume_calculation/no_vol.i)
- (tutorials/msfr/openmc.i)
- (test/tests/neutronics/feedback/multi_component_density/openmc.i)
- (test/tests/neutronics/relaxation/cell_tallies/openmc_nonaligned.i)
- (test/tests/neutronics/feedback/temperature/one_one/openmc.i)
- (test/tests/neutronics/feedback/multi_component_density/openmc_incorrect_length.i)
- (test/tests/neutronics/filters/sh/openmc_l1.i)
- (test/tests/openmc_errors/mesh_tally/incorrect_scaling.i)
- (test/tests/neutronics/tally_system/multi_cell_relax.i)
- (test/tests/openmc_errors/mode/plot/plot.i)
- (test/tests/openmc_errors/cell_instances/insufficient_materials/solid.i)
- (test/tests/neutronics/dagmc/mesh_tallies/csg_step_1/openmc.i)
- (test/tests/neutronics/filters/multi_filter.i)
- (test/tests/neutronics/source/openmc.i)
- (test/tests/neutronics/kinetics/both.i)
- (test/tests/neutronics/filters/filter_no_norm_score.i)
- (test/tests/neutronics/heat_source/default_tally_blocks.i)
- (test/tests/neutronics/feedback/multi_component_density/multi/multi_vars_alt.i)
- (test/tests/neutronics/dagmc/cell_tallies/csg_step_2/openmc.i)
- (test/tests/neutronics/mesh_tally/one_mesh_no_input_file.i)
- (tutorials/gas_compact_multiphysics/openmc_nek.i)
- (test/tests/neutronics/tally_system/multi_mesh_relax.i)
- (test/tests/neutronics/feedback/unmapped_moose/openmc.i)
- (test/tests/openmc_errors/input_params/levels.i)
- (test/tests/neutronics/heat_source/partial_overlap_moose_union.i)
- (test/tests/openmc_errors/xs_temperatures/openmc.i)
- (test/tests/neutronics/feedback/triso/different_fill_univs/openmc.i)
- (test/tests/neutronics/mesh_tally/block_restrict.i)
- (test/tests/neutronics/symmetry/triso/openmc.i)
- (test/tests/openmc_errors/mode/particle/particle.i)
- (test/tests/openmc_errors/tallies/length.i)
- (tutorials/pincell_multiphysics/openmc.i)
- (test/tests/neutronics/dagmc/incompatible_geom/mismatch/openmc.i)
- (test/tests/neutronics/relaxation/mesh_tallies/output_fission_tally.i)
- (tutorials/lwr_solid/openmc.i)
- (tutorials/lwr_amr/openmc_amr_single.i)
- (tutorials/lwr_solid/openmc_um.i)
- (test/tests/neutronics/heat_source/from_postprocessor.i)
- (test/tests/openmc_errors/mesh_tally/incorrect_file.i)
- (test/tests/neutronics/mesh_tally/one_mesh_global.i)
- (test/tests/openmc_errors/input_params/invalid_max_batches.i)
- (test/tests/userobjects/volume_calculation/scaling.i)
- (test/tests/neutronics/tally_system/multi_cell_triggers.i)
- (test/tests/openmc_errors/level/total_too_high/level_too_high.i)
- (test/tests/neutronics/feedback/interchangeable/no_tally.i)
- (test/tests/openmc_errors/input_params/no_dag.i)
- (tutorials/lwr_amr/openmc.i)
- (test/tests/neutronics/feedback/different_units/openmc.i)
- (test/tests/neutronics/feedback/triso/missing_triso_fill/openmc.i)
- (test/tests/neutronics/triggers/mesh_tally_rel_err.i)
- (test/tests/openmc_errors/material_mappings/materials.i)
- (test/tests/neutronics/heat_source/distrib_cell/solid.i)
- (test/tests/neutronics/moving_mesh/mesh_tally_file.i)
- (tutorials/gas_compact_multiphysics/openmc_thm.i)
- (test/tests/symmetry/rotation.i)
- (test/tests/neutronics/dagmc/mesh_tallies/with_scaling.i)
- (test/tests/neutronics/gen_mgxs/mgxs_aux/generic_mgxs.i)
- (test/tests/neutronics/feedback/multi_component_temp/multi_vars.i)
- (doc/content/source/problems/smallest_openmc_input.i)
- (test/tests/neutronics/dagmc/mesh_tallies/csg_step_2/openmc.i)
- (test/tests/neutronics/filters/sh/openmc_l1_mult.i)
- (test/tests/neutronics/adaptivity/unchanged_sub.i)
- (test/tests/neutronics/gen_mgxs/mgxs_aux/mg_diffusion_coeff.i)
- (test/tests/neutronics/photon/openmc.i)
- (test/tests/neutronics/heat_source/multi_tally_overlap_solid.i)
- (test/tests/neutronics/dagmc/properties.i)
- (test/tests/neutronics/tally_system/multi_cell.i)
- (test/tests/openmc_errors/block_mappings/absent_solid_block.i)
- (test/tests/neutronics/dagmc/mesh_tallies/disjoint_bins.i)
- (test/tests/neutronics/filters/azimuthal/mesh.i)
- (test/tests/postprocessors/openmc_particles/openmc.i)
- (test/tests/neutronics/solid/openmc_zero.i)
- (test/tests/neutronics/indicators/rel_err/openmc.i)
- (test/tests/neutronics/feedback/temperature/one_two/openmc.i)
- (test/tests/neutronics/filters/polar/mesh_sum.i)
- (test/tests/neutronics/tally_system/multi_mesh_triggers.i)
- (test/tests/neutronics/dagmc/with_csg/mixed_feedback/openmc.i)
- (test/tests/neutronics/filters/polar/cell.i)
- (test/tests/neutronics/openmc_xml/skip_statepoint.i)
- (test/tests/neutronics/filters/xml/mesh.i)
- (test/tests/neutronics/relaxation/cell_tallies/openmc.i)
- (test/tests/neutronics/feedback/different_units/openmc_cm.i)
- (test/tests/openmc_errors/fixed_source/k.i)
- (test/tests/neutronics/feedback/lattice/openmc.i)
- (test/tests/neutronics/feedback/interchangeable/openmc.i)
- (test/tests/neutronics/feedback/triso/openmc.i)
- (test/tests/postprocessors/reactivity/openmc.i)
- (test/tests/neutronics/gen_mgxs/all_mgxs_cell_l0.i)
- (test/tests/neutronics/heat_source/cell_volumes.i)
- (test/tests/neutronics/mesh_tally/different_units_and_translations.i)
- (test/tests/neutronics/feedback/multi_component_temp/openmc.i)
- (test/tests/neutronics/moving_mesh/cell_tally.i)
- (tutorials/pebbles/openmc.i)
- (test/tests/neutronics/mesh_tally/one_mesh.i)
- (test/tests/neutronics/moving_mesh/temperature.i)
- (test/tests/neutronics/mesh_tally/no_coupling.i)
- (test/tests/neutronics/filters/energy/mesh_sum.i)
- (test/tests/neutronics/moving_mesh/density.i)
- (test/tests/neutronics/triggers/tally_rel_err.i)
- (test/tests/neutronics/triggers/k_std_dev.i)
- (test/tests/openmc_errors/input_params/invalid_batches.i)
- (test/tests/neutronics/dagmc/cell_tallies/openmc.i)
- (test/tests/neutronics/gen_mgxs/all_mgxs_mesh.i)
- (test/tests/userobjects/openmc_nuclide_densities/only_density.i)
- (test/tests/neutronics/output/cell.i)
- (test/tests/neutronics/dagmc/cell_tallies/unmapped_cells.i)
- (test/tests/neutronics/feedback/multiple_levels/openmc.i)
- (test/tests/neutronics/feedback/materials/openmc.i)
- (test/tests/neutronics/feedback/temperature/two_one/openmc_multi_temp.i)
- (test/tests/neutronics/dagmc/with_csg/dag_lattice_outer/openmc.i)
- (test/tests/openmc_errors/densities/void_density.i)
- (tutorials/pebbles/openmc_um.i)
- (test/tests/controls/openmc_nuclide_densities/openmc.i)
- (test/tests/openmc_errors/level/phase_too_high/fluid_too_high.i)
- (test/tests/openmc_errors/mesh_translations/invalid_row.i)
- (test/tests/neutronics/triggers/multi_rel_err.i)
- (test/tests/neutronics/dagmc/missing_graveyard.i)
- (test/tests/neutronics/relaxation/cell_tallies/multi_tally.i)
- (test/tests/neutronics/dagmc/with_csg/openmc.i)
- (test/tests/neutronics/gen_mgxs/photon_xs/mgxs_photon.i)
- (test/tests/openmc_errors/cell_instances/lattice_outer/temp.i)
- (test/tests/neutronics/solid/openmc_missing_pebble.i)
- (test/tests/neutronics/symmetry/openmc.i)
- (test/tests/postprocessors/fission_tally_relative_error/ratio.i)
- (test/tests/neutronics/heat_source/multi_tally.i)
- (test/tests/openmc_errors/block_mappings/multiple_tally_settings.i)
- (test/tests/neutronics/moving_mesh/scale.i)
- (test/tests/openmc_errors/densities/zero_density.i)
- (test/tests/neutronics/filters/sh/openmc_l0_from_l1.i)
- (test/tests/neutronics/feedback/temperature/two_one/openmc.i)
- (test/tests/neutronics/output/mesh.i)
- (test/tests/neutronics/filters/energy_out/cell.i)
- (test/tests/neutronics/heat_source/overlap_solid.i)
- (test/tests/neutronics/dagmc/with_csg/multiple_dag/openmc.i)
- (test/tests/neutronics/filters/mesh_sum_gold.i)
- (test/tests/neutronics/adaptivity/unchanged_mesh.i)
- (test/tests/neutronics/markers/look_ahead/openmc.i)
- (tutorials/gas_compact/openmc.i)
- (test/tests/neutronics/heat_source/partial_overlap_openmc_union.i)
- (tutorials/dagmc/openmc.i)
- (test/tests/neutronics/tally_grad/finite_diff/openmc.i)
- (test/tests/userobjects/openmc_nuclide_densities/thermal_density.i)
- (test/tests/neutronics/moving_mesh/volumes.i)
- (test/tests/neutronics/adaptivity/mesh.i)
- (test/tests/neutronics/moving_mesh/mesh_tally.i)
- (test/tests/neutronics/adaptivity/cell.i)
- (test/tests/neutronics/dagmc/cell_tallies/one_bin.i)
- (test/tests/neutronics/heat_source/overlap_all.i)
- (test/tests/neutronics/filters/xml/cell.i)
- (test/tests/neutronics/relaxation/cell_tallies/dufek_gudowski.i)
- (test/tests/neutronics/dagmc/wrong_uo.i)
- (test/tests/neutronics/mesh_tally/multiple_meshes_global.i)
- (test/tests/postprocessors/fission_tally_relative_error/multi.i)
- (test/tests/userobjects/openmc_tally_editor/openmc_base.i)
- (test/tests/neutronics/dagmc/cell_tallies/null_density.i)
- (tutorials/csg_to_cad/openmc.i)
- (test/tests/neutronics/feedback/multi_component_density/multi/multi_vars.i)
- (test/tests/neutronics/mesh_tally/different_units.i)
- (tutorials/lwr_amr/openmc_amr.i)
- (test/tests/userobjects/openmc_nuclide_densities/openmc.i)
- (tutorials/gas_assembly/openmc.i)
- (test/tests/neutronics/dagmc/cell_tallies/csg_step_1/openmc.i)
- (test/tests/openmc_errors/tallies/separate_tallies.i)
- (test/tests/neutronics/filters/no_filter.i)
- (test/tests/neutronics/relaxation/cell_tallies/openmc_adapt.i)
- (test/tests/postprocessors/coupled_cells/openmc.i)
- (test/tests/openmc_errors/input_params/missing_properties.i)
- (test/tests/neutronics/gen_mgxs/subset_cell.i)
- (test/tests/openmc_errors/block_mappings/nonexistent_block.i)
- (test/tests/neutronics/flux/missing_norm.i)
- (test/tests/postprocessors/eigenvalue/openmc.i)
- (test/tests/openmc_errors/cell_instances/insufficient_materials/fluid.i)
- (test/tests/neutronics/gen_mgxs/hide_tally_vars.i)
- (test/tests/neutronics/tally_system/multi_estimator.i)
- (test/tests/neutronics/filters/energy/cell.i)
- (tutorials/tokamak/openmc.i)
- (test/tests/neutronics/feedback/lattice/non_material_fluid.i)
- (test/tests/neutronics/feedback/triso/cache/openmc.i)
- (test/tests/neutronics/dagmc/density_skin/csg_step_2/openmc.i)
- (test/tests/openmc_errors/mode/volume/volume.i)
- (test/tests/neutronics/filters/azimuthal/mesh_sum.i)
- (test/tests/neutronics/dagmc/density_skin/disjoint_bins.i)
- (test/tests/neutronics/dagmc/cell_tallies/with_scaling.i)
- (test/tests/neutronics/tally_system/multi_mesh.i)
- (test/tests/neutronics/gen_mgxs/all_mgxs_cell.i)
- (test/tests/neutronics/flux/flux.i)
- (test/tests/userobjects/volume_calculation/user_bb.i)
- (test/tests/neutronics/dagmc/density_skin/openmc.i)
- (test/tests/neutronics/relaxation/cell_tallies/output_fission_tally.i)
- (test/tests/neutronics/gen_mgxs/mgxs_aux/mg_tc_scatter_xs.i)
- (test/tests/userobjects/volume_calculation/openmc.i)
- (test/tests/openmc_errors/zero_tally/openmc.i)
- (test/tests/neutronics/filters/legendre/cell_l0_from_l1.i)
- (test/tests/neutronics/fixed_source/flux.i)
- (test/tests/neutronics/kinetics/beta.i)
- (test/tests/neutronics/filters/energy/mesh.i)
- (test/tests/neutronics/relaxation/mesh_tallies/openmc.i)
- (test/tests/neutronics/feedback/temperature/two_one/openmc_default.i)
- (test/tests/postprocessors/fission_tally_relative_error/openmc.i)
- (test/tests/neutronics/dagmc/no_symmetry.i)
- (test/tests/neutronics/density/openmc.i)
- (doc/content/source/actions/multi_tally_example.i)
- (test/tests/neutronics/feedback/lattice/openmc_scores.i)
- (test/tests/neutronics/dagmc/with_csg/dag_lattice/openmc.i)
- (test/tests/neutronics/heat_source/overlap_fluid.i)
- (test/tests/controls/openmc_nuclide_densities/error.i)
- (test/tests/neutronics/dagmc/density_skin/only_solid.i)