Multi-Group Cross Section Generation
In this tutorial, you will learn how to generate Multi-Group Cross Sections (MGXS) with distributed cell tallies to couple Cardinal with deterministic transport codes. To access this tutorial,
In this tutorial, we assume the user is familiar with mesh generation via the Reactor module and running LWR calculations with Cardinal-OpenMC. The LWR AMR tutorial is used as a base for this tutorial; we recommend the LWR AMR tutorial for users not familiar with these concepts.
Geometry and Computational Models
This model consists of a single UO assembly from the 3D C5G7 extension case in Smith et al. (2006), where control rods are fully inserted in the assembly to induce strong axial and radial gradients. Instead of using the multi-group cross sections from the C5G7 benchmark specifications, the material properties in Cathalau et al. (1996) are used. At a high level the geometry consists of the following lattice elements in a 17x17 grid:
264 fuel pins composed of UO pellets clad in zirconium with a helium gap;
24 control rods composed of BC pellets clad in aluminum, with no gap, occupying the assembly guide tubes;
A single fission chamber in the central guide tube composed of borated water, with a trace amount of U-235, clad in aluminum with no gap.
The remainder of the assembly not filled with these pincells is composed of borated water. Above the top of the fuel, there is a reflector region which is penetrated by the inserted control rods. The relevant dimensions can be found in Table 1.
Table 1: Geometric specifications for a rodded LWR assembly
| Parameter | Value (cm) |
|---|---|
| Fuel pellet outer radius | 0.4095 |
| Fuel clad inner radius | 0.418 |
| Fuel clad outer radius | 0.475 |
| Control rod outer radius | 0.3400 |
| Fission chamber outer radius | 0.3400 |
| Guide tube clad outer radius | 0.54 |
| Pin pitch | 1.26 |
| Fuel height | 192.78 |
| Reflector thickness | 21.42 |
When generating MGXS, tallies do not need to be normalized to power, but Cardinal still requires a value of reactor power when performing k-eigenvalue calculations. For this tutorial we select an arbitrary power of 1 Wth. If you want to compute cross sections accounting for multi-physics feedback, a true reactor power and a coupled thermal-hydraulics solver are required.
OpenMC Model
The OpenMC model follows standard model building practices for LWR geometries. The Python script used to generate the model.xml file can be found below, and a plot of the geometry can be found in Figure 1.
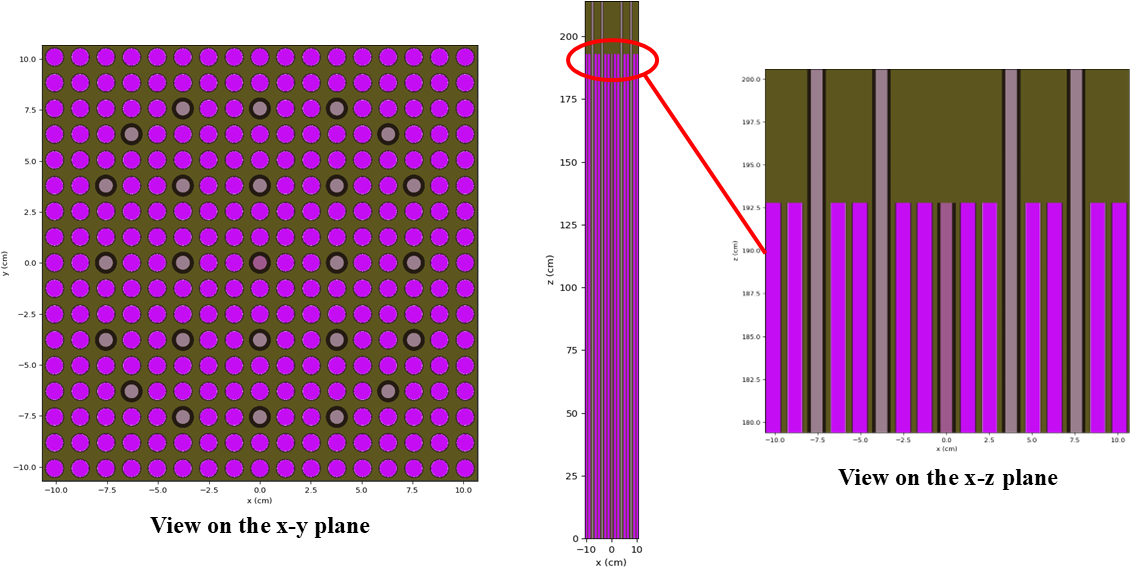
Figure 1: OpenMC geometry colored by material ID shown on the - and - planes
To generate the XML files needed to run OpenMC, you can run the following:
Or you can use the model.xml file that is included in the tutorials/lwr_mgxs directory.
Mesh Mirror for MGXS Generation
Most MGXS workflows store the resulting cross sections in custom libraries for specific deterministic transport codes. Instead, Cardinal determines homogenization volumes using the mesh mirror, and stores the cross sections on the mesh at the end of the OpenMC simulation. This process allows for "transfering" of MGXS between Cardinal and a coupled deterministic code using the multi-app system. This determininstic code coupling enables higher fidelity cross sections generation and recalculation of group properties to account for multi-physics feedback. The input file used to generate this mesh mirror can be found below.
(tutorials/lwr_mgxs/mesh.i)The mesh is similar to the mesh generated in the AMR tutorial with a few key differences to accomodate a coupled deterministic transport code. We set NUM_SECTORS and FUEL_RADIAL_DIVISIONS to 4 to increase the radial fidelity of of each pin. The increased discretization is necessary for deterministic calculations as these regions have strong gradients due to absorption in the fuel and control rods. We also avoid deleting the gap blocks and extruded the mesh an additional 21.42 cm to cover the reflector. These changes are necessary to preserve mesh connectivity for deterministic transport solvers using these cross sections, and to ensure we generate cross sections for the reflector above the core.
The input mesh Cardinal will use to store cell tally results on (mesh_in.e) can be generated by running the command below. The resulting mesh can be found in Figure 2; we note that this mesh is substantially finer in then a conventional Cardinal mesh mirror to adequately capture spatial gradients in the determistic solver.

Figure 2: The mesh mirror used for computing and storing MGXS, shown with an isometric view and sliced on the plane
Neutronics Input File
The neutronics calculation is performed over the entire assembly by OpenMC and the wrapping of the OpenMC results in MOOSE is performed in openmc_mgxs.i. We begin by defining the mesh (generated in the previous step) used by Cardinal to determine which cell tallies should be constructed for MGXS generation:
Afterwards, we add two auxvariables and auxkernels for post-processing the cross sections – we use these to compute the scattering ratio (the sum of scattering cross sections divided by the total cross section) in each energy group. The scattering ratio for each group should be less than or equal to one; scattering ratios larger than one may result in non-convergence of the deterministic solver or convergence to incorrect results. Note that we did not delete the gap blocks in the mesh, but we are block restricting the auxkernels and auxvariables such that they are not computed over the gap region (block 2). This is necessary as the gaps do not receive any tally hits in OpenMC. Therefore, the cross sections will be zero resulting in a divide by zero when computing scattering ratios.
(tutorials/lwr_mgxs/openmc_mgxs.i)Next, the Problem block (with an OpenMCCellAverageProblem) describes the syntax necessary to replace the normal MOOSE finite element calculation with an OpenMC neutronics solve. We select cell_level = 1 to ensure that we generate our cell to element mapping for an OpenMC geometry depth one lower than the root universe. This will result in a distributed cell tally for each unique geometric region in the LWR lattice, as opposed to the entire lattice (which is what would happen with cell_level = 0). We then specify the number of particles (particles = 10000), the number of inactive batches (inactive_batches = 50), and the total number of batches (batches = 1000).
After specifying the basis problem settings, we move on to setting up the MGXS parameters with the MGXS block. We specify a distributed cell tally should be used as the MGXS homogenization domain with tally_type = cell and the CASMO 2 group structure as the energy group boundaries. We then select particle = neutron to set the particle filter to use when computing cross sections, which is useful when OpenMC is performing coupled neutron-photon transport. We then specify estimator = 'analog', which is required when computing scattering cross sections and the fission spectra. For the same reasons as the auxkernels added for post-processing, we block restrict the MGXS such that they are not computed on the gap region. Finally, we select the MGXS that we wish to compute. This includes total cross sections (always computed), scattering matrix cross sections (enabled by default), nu-fission cross sections (add_fission = true), fission chi spectra (add_fission = true), and fission heating cross sections (add_fission_heating = true). Note, other cross sections or group properties can be computed; an exhaustive list of the options can be found in the documentation for the MGXS block. We choose to disable the transport correction (transport_correction = false) for the scattering cross sections to avoid pushing the scattering ratio over unity, which can occur when there aren't enough particles scoring to higher order scattering moments.
Since we've selected add_fission_heating = true, we don't need to add other heating tallies for normalization purposes – we can set source_rate_normalization = 'kappa_fission' in the Problem block. A steady-state executioner is selected as OpenMC will run a single criticality calculation. We then select exodus output which will occur on the end of the simulation.
Execution and Postprocessing
To run the wrapped neutronics calculation,
This will run OpenMC with 2 MPI ranks with 4 OpenMP threads per rank. To run the simulation faster, you can increase the parallel processes/threads, or simply decrease the number of particles used in OpenMC. When the simulation is complete, you will have created openmc_out.e, an Exodus mesh with the computed cross sections. In the console output, you should see the following tally list:
Each of these tallies are automatically applied to the cells discovered by Cardinal when generating the cell to element mapping. The generated cross sections for the fuel can be found in Figure 3, where we can see the importance of accurately capturing spatial and spectral effects when generating group constants. The fast neutron production cross sections for the corner pins of the LWR assembly are larger then the central pins due to spatial shielding effects. The thermal neutron production cross section increases in the region between the center and periphery of the assembly due to an increase in moderation. Additionally, a depression in the thermal neutron production cross section can also be seen in the fuel pins close to the inserted control rods.
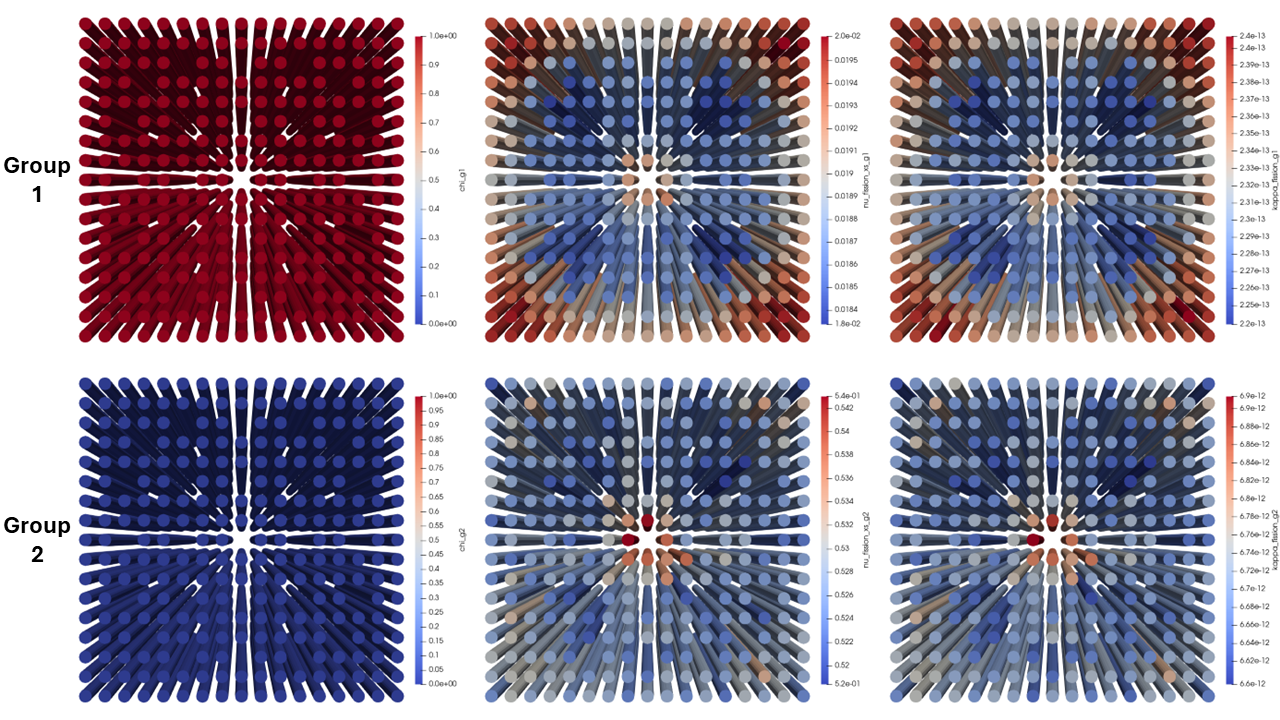
Figure 3: The generated MGXS at the core centerline. Left: . Middle: . Right: .
The group-wise total cross section and scattering ratio for all materials at the core centerline can be found in Figure 4. Spatial trends are difficult to pick out due to the order-of-magnitude difference between MGXS for the different materials. However, we see the control rods are the strongest absorbers in both the fast and thermal range. The borated water coolant has a very large scattering ratio in both the fast and thermal groups, showcasing the effectiveness of water as a neutron moderator.

Figure 4: The generated MGXS at the core centerline. Left: . Right: .
While not a capability available in Cardinal, we take these MGXS and use them as an input in a deterministic solver to showcase the end result of a standard two-step calculations. The fast and thermal fluxes from the discrete-ordinates code Gnat (Sawatzky and Atkinson (2024)) using these cross sections can be found in Figure 5 and Figure 6. The predicted continuous-energy value of from Cardinal-OpenMC is . Gnat (using 200 directions per energy group) obtains a multi-group of , yielding pcm. There is likely some amount of cancelation of error in the deterministic result, evidenced by the surprisingly good agreement obtained without using transport-corrected scattering cross sections. However, we believe these results showcase the value of this automated workflow for computing spatially-varying cross sections for deterministic transport calculations.
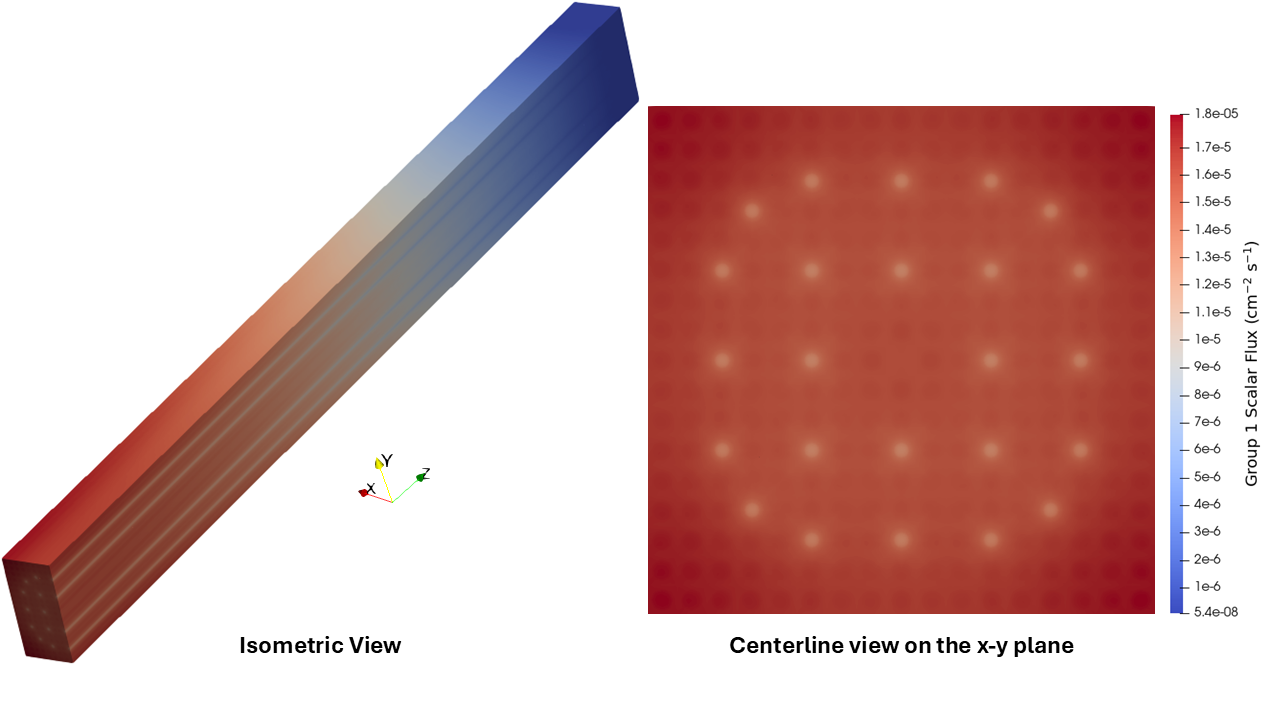
Figure 5: Group 1 (fast) fluxes from Gnat using the generated MGXS. Fluxes are normalized such that the integral over all space and energy is unity.

Figure 6: Group 2 (thermal) fluxes from Gnat using the generated MGXS. Fluxes are normalized such that the integral over all space and energy is unity.
References
- S. Cathalau, J. C. Lefebvre, and J. P. West.
Proposal for a Second Stage of the Benchmark on Power Distributions within Assemblies.
Technical Report NEA/NSC/DOC(96)2, Organisation for Economic Cooperation and Development, Nuclear Energy Agency, 1996.
URL: https://www.oecd-nea.org/upload/docs/application/pdf/2020-01/nsc-doc96-02-rev2.pdf.[BibTeX]
- K. Sawatzky and K. D. Atkinson.
Verification of a CARIBOU-OpenMC Workflow for the Analysis of HTGR-Like Systems Using the Proposed Ontario Tech Subcritical Assembly.
In Proceedings of PHYSOR. 2024.[BibTeX]
- M.A. Smith, E.E. Lewis, and Byung-Chan Na.
Benchmark on Deterministic 3-D MOX Fuel Assembly Transport Calculations without Spatial Homogenization.
Progress in Nuclear Energy, 48(5):383–393, 2006.
doi:https://doi.org/10.1016/j.pnucene.2006.01.002.[BibTeX]