Multiphysics for a Molten Salt Fast Reactor
In this tutorial, you will learn how to:
Couple OpenMC and NekRS for modeling a Molten Salt Fast Reactor (MSFR)
Use DAGMC Computer Aided Design (CAD) geometry in OpeNMC
Use on-the-fly geometry adaptivity ("skinning") to change the OpenMC cells in response to multiphysics feedback
To access this tutorial,
This tutorial also requires you to download some mesh and restart files from Box. Please download the files from the msfr folder here.
This tutorial requires High Performance Computing (HPC) for running the NekRS model. You will be able to run the OpenMC model without HPC resources. Note that you need to have built Cardinal with DAGMC support enabled, by setting export ENABLE_DAGMC=true.
Geometry and Computational Model
The geometry consists of an MSFR with geometry from Rouch et al. (2014). Volume slices through the NekRS and OpenMC domains are shown in Figure 1.
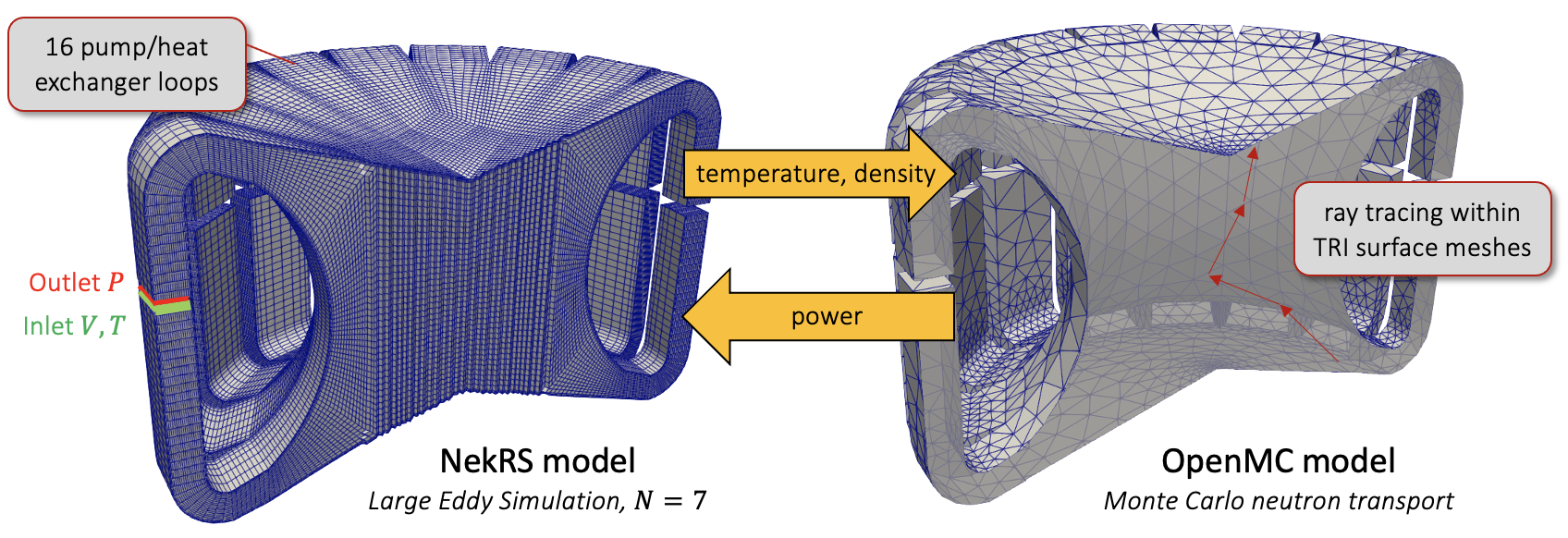
Figure 1: Volume slices through the NekRS and OpenMC domains
As this tutorial is intended primarily to demonstrate on-the-fly geometry skinning, several important features are neglected such as the transport of Delayed Neutron Precursors (DNPs). Future extensions will incorporate material movement in OpenMC driven by velocity (e.g. from a CFD solver such as NekRS), which allows this assumption to be relaxed in the future. For geometric simplicity, radial breeder blankets and axial reflectors are also neglected. The OpenMC model consists of the triangulated surface in Figure 1, with a larger bounding box surrounding it which is set to a vacuum boundary condition.
The nominal operating conditions, as well as the actual conditions used in this tutorial, are summarized in Table 1. In the present tutorial, the reactor is modeled at a 10% flow, 10% power condition (i.e. 300 MWth and 1893 kg/s) to be within an approachable range for Large Eddy Simulation (LES). The NekRS model uses LES at polynomial oder (512 degrees of freedom per element) for 616000 hexahedral elements, or 315 million grid points. Note in Figure 1 that the meshes used by NekRS and OpenMC are very different - the OpenMC domain uses a triangulated mesh, with about 35000 elements.
Table 1: Nominal and simulated operating conditions for the MSFR
| Parameter | Nominal Value | Simulated Value |
|---|---|---|
| Inlet temperature (K) | 898 | 898 |
| Bulk outlet temperature (K) | 998 | 998 |
| Power (MWth) | 3000 | 300 |
| Mass flowrate (kg/s) | 18923 | 1893 |
The heat exchanger and pump in each of the 16 legs are replaced by recycling inlet and turbulent outlet boundary conditions for velocity. The recycling inlet boundary condition applies the inlet velocity using the velocity distribution some distance upstream from the outlet boundary, essentially representing the inlet as fully-developed channel flow. The inlet temperature is then set to a uniform value of 898 K. For simplicity, the NekRS model uses an incompressible flow model with constant thermophysical properties from Rouch et al. (2014). A Boussinesq buoyancy term will be added in future work. In the OpenMC model, density feedback is approximated by evaluating a thermophysical property correlation given temperature feedback from NekRS.
OpenMC Model
OpenMC is used to solve for the neutron transport and power distribution. The OpenMC model uses DAGMC for ray tracing within triangular surface meshes in -eigenvalue mode. The OpenMC model is created using OpenMC's Python API. As shown in Figure 1, the initial OpenMC model consists of a single large volume, enclosed by one large triangulated mesh surface. This geometry will be adaptively updated according to multiphysics feedback.
The OpenMC model is created with the model.py script. The geometry is created from a triangulated volume mesh, exported from Cubit. We create a single material to represent the salt - notice how this contrasts with easlier tutorials for Constructive Solid Geometry (CSG) geometries, where we needed to make a unique material for every unique density region. Later, Cardinal will automatically create new materials in memory when we skin the geometry.
The initial DAGMC model does not contain a graveyard body - so you cannot run this file in a standalone OpenMC run. Cardinal will create a graveyard for you when you have skinning enabled.
You can create these XML files by running
or you can simply use the XML files versioned in the directory.
NekRS Model
NekRS is used to solve the incompressible Navier-Stokes equations in non-dimensional form. The NekRS input files needed to solve the incompressible Navier-Stokes equations are:
msfr.re2: NekRS meshmsfr.par: High-level settings for the solver, boundary condition mappings to sidesets, and the equations to solvemsfr.udf: User-defined C++ functions for on-line postprocessing and model setupmsfr.oudf: User-defined Open Concurrent Compute Abstraction (OCCA) kernels for boundary conditions and source termsmsfr.usr: User-defined Fortran functions; these are used to apply the recycling boundary condition. Long term, eventually all Nek5000 capabilities will be ported to NekRS, and this fortran file would not be necessary.
A detailed description of all of the available parameters, settings, and use cases for these input files is available on the NekRS documentation website. Because the purpose of this analysis is to demonstrate Cardinal's capabilities, only the aspects of NekRS required to understand the present case will be covered.
The mesh is created offline using gmsh, so we simply provide the final mesh on Box. Next, the .par file contains problem setup information. This input solves for pressure, velocity, and temperature. In the nondimensional formulation, the "viscosity" becomes , where is the Reynolds number, while the "thermal conductivity" becomes , where is the Peclet number. These nondimensional numbers are used to set various diffusion coefficients in the governing equations with syntax like -4.8e4, which is equivalent in NekRS syntax to .
We use the high-pass filter in NekRS for the LES, and filter the highest two modes. We set a Reynolds number of and a Prandtl number of 17.05. We restart this simulation from a previous standalone NekRS run, which used a "frozen" power distribution given in the literature Rouch et al. (2014).
(tutorials/msfr/msfr.par)Next, the .udf file is used to set up a heat source GPU kernel which we will use to send OpenMC's heat source from Cardinal into NekRS. As we will show later, this heat source is occupying the first (0-indexed) slot of the scratch space. In the UDF_ExecuteStep, we also do some special copies from some Nek5000 backend data (special usage here to accomplish the recycling boundary conditions) into the 3rd, 4th, and 5th slots of the scratch space (the scratch space is zero-indexed). As shown in the .par file, we will read initial conditions for velocity, pressure, and temperature from a restart file, restart.fld, so we don't need to set any other initial conditions here.
In the .oudf file, we define boundary conditions and any GPU kernels. We see in velocityDirichletConditions how we are setting the inlet velocities equal to the 3rd, 4th, and 5th slots in the scratch space. The mooseHeatSource is a GPU kernel simply reading from scratch space into the QVOL array (which holds a user-defined volumetric heating).
Finally, we have a .usr file with Fortran code which accesses from features in Nek5000 which have not yet been fully ported over to NekRS. The code in this file is being used to apply the recycling boundary conditions.
Multiphysics Coupling
In this section, OpenMC and NekRS are coupled for multiphysics modeling of an MSFR. This section describes all input files.
OpenMC Input Files
The neutron transport is solved using OpenMC. The input file for this portion of the physics is openmc.i. We begin by setting up the mesh mirror, which is the same volumetric mesh used to generate the DAGMC geometry.
Next, we set some initial conditions for temperature and density, because we will run OpenMC first.
(tutorials/msfr/openmc.i)Next, we define a number of auxiliary variables to be used for diagnostic purposes. With the exception of the ParsedAux used to compute a fluid density in terms of temperature, none of the following variables are necessary for coupling, but they will allow us to visualize how data is mapped from OpenMC to the mesh mirror. The CellTemperatureAux and CellDensityAux will display the OpenMC cell temperatures and densities (after volume-averaging from Cardinal).
(tutorials/msfr/openmc.i)Next, the [Problem] and [Tallies] blocks define all the parameters related to coupling OpenMC to MOOSE. We will send temperature and density to OpenMC, and extract power using a MeshTally. We set a number of relaxation settings to use Dufek-Gudowski relaxation, which will progressively ramp the number of particles used in the simulation (starting at 5000) so that we selectively apply computational effort only after the thermal-fluid physics are reasonably well converged. Finally, we will be "skinning" the geometry on-the-fly by providing the skinner user object (of type MoabSkinner).
After every Picard iteration, the skinner will group the elements in the mesh according to their temperature and density. For this tutorial, we will group the elements according to temperature by 15 different bins, ranging from a minimum temperature of 800 K up to a maximum temperature of 1150 K. We will then also bin by density, so that we will have a second grouping according to density. Then, we will create a new OpenMC cell for each unique combination of temperature and density, and re-generate the DAGMC mesh surfaces that bound these regions.
Next, we create a NekRS sub-application, and set up transfers of data between OpenMC and NekRS. These transfers will send fluid temperature and density from NekRS up to OpenMC, and a power distribution to NekRS. We will use sub-cycling, and only send data to/from NekRS at those synchronization points, by using the synchronize_in postprocessor transfer.
Finally, we will use a Transient executioner and add a number of postprocessors for diagnostic purposes. We set the "time step" size in OpenMC to be equal to 2000 times the (dimensional) NekRS time step size, so we are essentially running 2000 NekRS time steps for each OpenMC -eigenvalue solve.
(tutorials/msfr/openmc.i)Fluid Input Files
The fluid mass, momentum, and energy transport physics are solved using NekRS. The input file for this portion of the physics is nek.i. We begin by defining a number of file-local constants and by setting up the NekRSMesh mesh mirror. Because we are coupling NekRS via volumetric heating to OepNMC, we need to use a volumetric mesh mirror. The characteristic length chosen for the NekRS files is already 1 m, so we do not need to scale the mesh in any way.
The bulk of the NekRS wrapping is specified with NekRSProblem. The NekRS input files are in non-dimensional form, whereas all other coupled applications use dimensional units. The various *_ref and *_0 parameters define the characteristic scales that were used to non-dimensionalize the NekRS input.
Then, we simply set up a Transient executioner with the NekTimeStepper.
(tutorials/msfr/nek.i)Execution
To run the input files,
This will produce a number of output files,
openmc_out.e, OpenMC simulation resultsopenmc_out_nek0.e, NekRS simulation results, mapped to aSECONDorder Lagrange basismoab_skins_*.h5m, OpenMC cell surfaces (in units of centimeters), which can be converted to.vtkformat using thembconvertscriptmsfr0.f*, NekRS output files
Figure 2 shows the OpenMC power (on a mesh tally), NekRS fluid temperature, and the OpenMC cell temperatures for the last Picard iteration. The black lines delineate the edges of the OpenMC cells, which have been adaptively generated according to the multiphysics feedback.
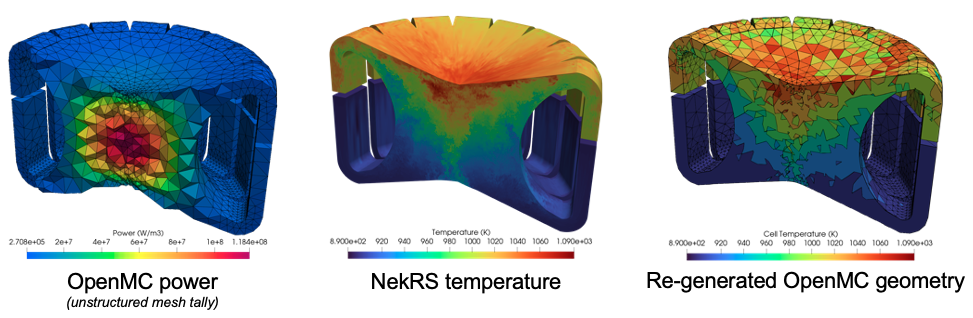
Figure 2: OpenMC power, NekRS fluid temperature, and re-generated OpenMC cells (delinated with black lines) showing the temperature in each cell
References
- H. Rouch, O. Geoffroy, P. Rubiolo, A. Laureau, M. Brovchenko, D. Heuer, and E. Merle-Lucotte.
Preliminary Thermal-Hydraulic Core Design of the Molten Salt Fast Reactor (MSFR).
Annals of Nuclear Energy, 64:449–456, 2014.
doi:10.1016/j.anucene.2013.09.012.[BibTeX]