When building an OpenMC model, there are several parameters that affect the convergence:
To ensure that a sufficient number of active batches are used, we recommend using the trigger system in the OpenMC wrapping, which will terminate the number of active batches once reaching a desired uncertainty in k and/or the kappa-fission tally. To determine an appropriate nuber of inactive batches and a cell discretization, Cardinal contains several helper scripts. This tutorial describes how to use these scripts.
Inactive Batches
To determine an appropriate number of inactive batches, you can use the scripts/inactive_study.py script. To run this script, you need:
A Python script that creates your OpenMC model for you; this Python script must take -s as a boolean argument that indicates a Shannon entropy mesh will be added, and -n <layers> to indicate the number of cell divisions to create
A Cardinal input file that runs your OpenMC model given some imposed temperature and density distribution
For example, to run an inactive cycle study for the model developed for the gas compact tutorial, we would run:
since the Python script used to generate the OpenMC model is named unit_cell.py and our Cardinal input file is named openmc.i. You should edit the beginning of the inactive_study.py script to select the number of cell layers and number of inactive cycles to consider:
use_saved_statepoints = False
n_layers = [5, 10]
n_inactive = 100
averaging_batches = 100
from argparse import ArgumentParser
import multiprocessing
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
import openmc
import sys
import os
matplotlib.rcParams.update({'font.size': 14})
colors = ['firebrick', 'orangered', 'darkorange', 'goldenrod', 'forestgreen', \
'lightseagreen', 'steelblue', 'slateblue']
program_description = ("Script for determining required number of inactive batches "
"for an OpenMC model divided into a number of layers")
ap = ArgumentParser(description=program_description)
ap.add_argument('-i', dest='script_name', type=str, required=True,
help='Name of the OpenMC python script to run (without .py extension)')
ap.add_argument('-n-threads', dest='n_threads', type=int,
default=multiprocessing.cpu_count(), help='Number of threads to run Cardinal with')
ap.add_argument('-input', dest='input_file', type=str, required=True,
help='Name of the Cardinal input file to run')
ap.add_argument('--method', dest = 'method', choices =['all','half','window','none'], default='none',
help = 'The method to estimate the number of sufficient inactive batches to discard before tallying in k-eigenvalue mode. ' +
'This number is determined by the first batch at which a running average of the Shannon Entropy falls within the standard ' +
'deviation of the run. Options are all, half, and window; all uses all batches, half uses the last half of the batches, and ' +
'window uses a user-specified number. Additionally, none can be specified if the feature is undesired.')
ap.add_argument('--window_length', dest = 'window_length', type = int,
help =' When the window method is selected, the window length must be specified. The window length is a number of batches '
' before the current batch to use when computing the running average and standard deviation.')
args = ap.parse_args()
method = args.method
if (args.method == 'window' and args.window_length == None):
raise TypeError('The method specified was window, but window_length = None. Please specify --window_length = LENGTH, where LENGTH is an integer')
else:
if (args.method == 'window'):
window_length = args.window_length
input_file = args.input_file
script_name = args.script_name + ".py"
file_path = os.path.realpath(__file__)
file_dir, file_name = os.path.split(file_path)
exec_dir, file_name = os.path.split(file_dir)
n_active = 10
methods = ['opt', 'devel', 'oprof', 'dbg']
exec_name = ''
for i in methods:
if (os.path.exists(exec_dir + "/cardinal-" + i)):
exec_name = exec_dir + "/cardinal-" + i
break
if (exec_name == ''):
raise ValueError("No Cardinal executable was found!")
if (not os.path.exists(os.getcwd() + '/inactive_study')):
os.makedirs(os.getcwd() + '/inactive_study')
entropy = []
k = []
k_generation_avg = []
n_batches = n_inactive + n_active
max_entropy = sys.float_info.min
min_entropy = sys.float_info.max
max_k = sys.float_info.min
min_k = sys.float_info.max
for n in n_layers:
new_sp_filename = 'inactive_study/statepoint_' + str(n) + '_layers.h5'
if ((not use_saved_statepoints) or (not os.path.exists(new_sp_filename))):
os.system("python " + script_name + " -s -n " + str(n))
os.system(exec_name + " -i " + input_file + \
" Problem/inactive_batches=" + str(n_inactive) + \
" Problem/batches=" + str(n_batches) + \
" Problem/max_batches=" + str(n_batches) + \
" MultiApps/active='' Transfers/active='' Executioner/num_steps=1" + \
" --n-threads=" + str(args.n_threads))
os.system('cp statepoint.' + str(n_batches) + '.h5 ' + new_sp_filename)
with openmc.StatePoint(new_sp_filename) as sp:
entropy.append(sp.entropy)
k.append(sp.k_generation)
max_entropy = max(np.max(sp.entropy[:n_inactive][:]), max_entropy)
min_entropy = min(np.min(sp.entropy[:n_inactive][:]), min_entropy)
max_k = max(np.max(sp.k_generation[:n_inactive][:]), max_k)
min_k = min(np.min(sp.k_generation[:n_inactive][:]), min_k)
averaging_k = sp.k_generation[(n_inactive - averaging_batches):n_inactive]
k_generation_avg.append(sum(averaging_k) / len(averaging_k))
entropy_range = max_entropy - min_entropy
for i in range(len(n_layers)):
nl = n_layers[i]
fig, ax = plt.subplots()
ax.plot(entropy[i][:n_inactive][:], label='{0:.0f} layers'.format(nl), color=colors[i])
ax.set_xlabel('Inactive Batch')
ax.set_ylabel('Shannon Entropy')
ax.set_ylim([min_entropy - 0.05 * entropy_range, max_entropy + 0.05 * entropy_range])
plt.grid()
plt.legend(loc='upper right')
plt.savefig('inactive_study/' + args.script_name + '_entropy' + str(nl) + '.pdf', bbox_inches='tight')
plt.close()
for i in range(len(n_layers)):
nl = n_layers[i]
fig, ax = plt.subplots()
ax.plot(k[i][:n_inactive][:], label='{0:.0f} layers'.format(nl), color=colors[i])
plt.axhline(y=k_generation_avg[i], color='k', linestyle='-')
ax.set_xlabel('Inactive Batch')
ax.set_ylabel('$k$')
ax.set_ylim([min_k, max_k])
plt.grid()
plt.legend(loc='lower right')
plt.savefig('inactive_study/' + args.script_name + '_k' + str(nl) + '.pdf', bbox_inches='tight')
plt.close()
if (method != 'none'):
for i in range(len(n_layers)):
nl = n_layers[i]
print("\nLayers: ", nl)
print("----------------------")
start = 1
if (method == "window"):
start = window_length
for j in range(start, len(entropy[i])):
if (method == "window"):
window = entropy[i][(j - window_length):j]
elif (method == "half"):
idx = int(np.floor(j/2))
window = entropy[i][idx:j]
else:
window = entropy[i][0:j]
window_mean = np.average(window)
window_dev = np.std(window)
window_low = window_mean - window_dev
window_high = window_mean + window_dev
extra_str = " "
if (entropy[i][j] <= window_high and entropy[i][j] >= window_low):
extra_str = "--> "
print(extra_str + "Inactive batch: {:6d} Entropy: {:.6f} Window mean: {:.6f} +/- {:.6f}".format(j, entropy[i][j], window_mean, window_dev))
print("--> indicates batch which satisfies method. DOES NOT necessarily indicate a converged fission source.")
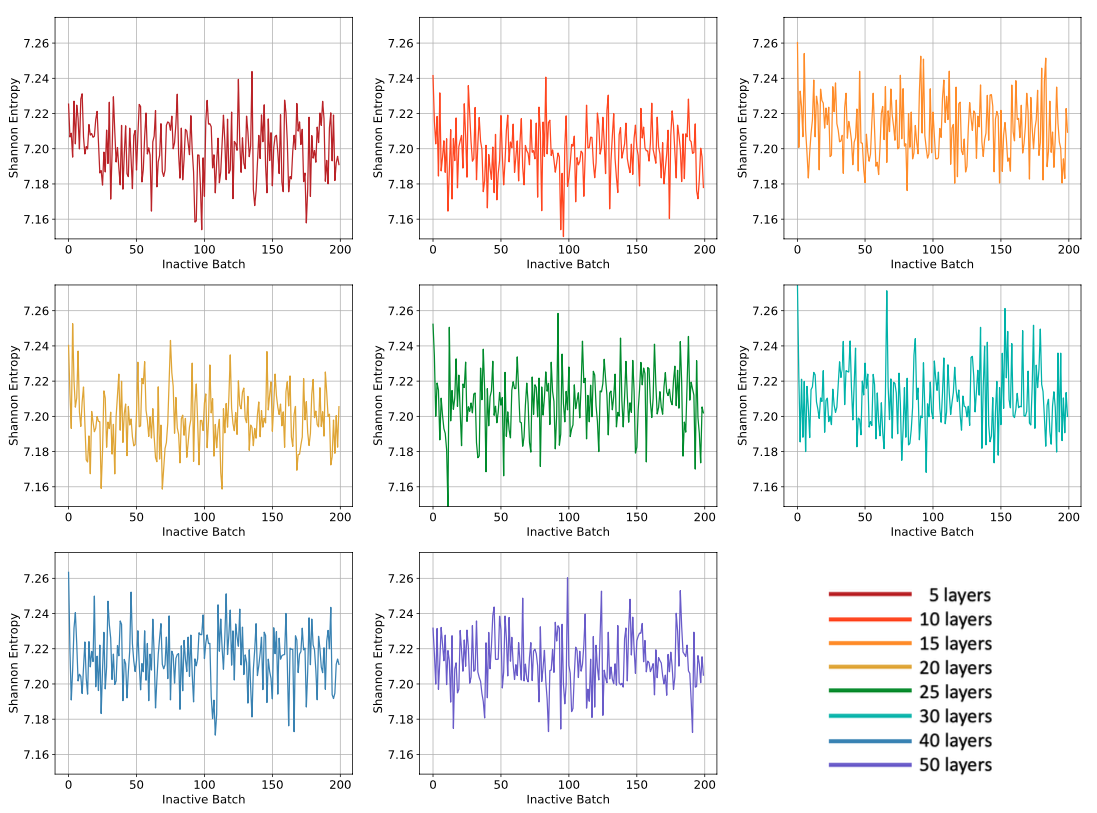
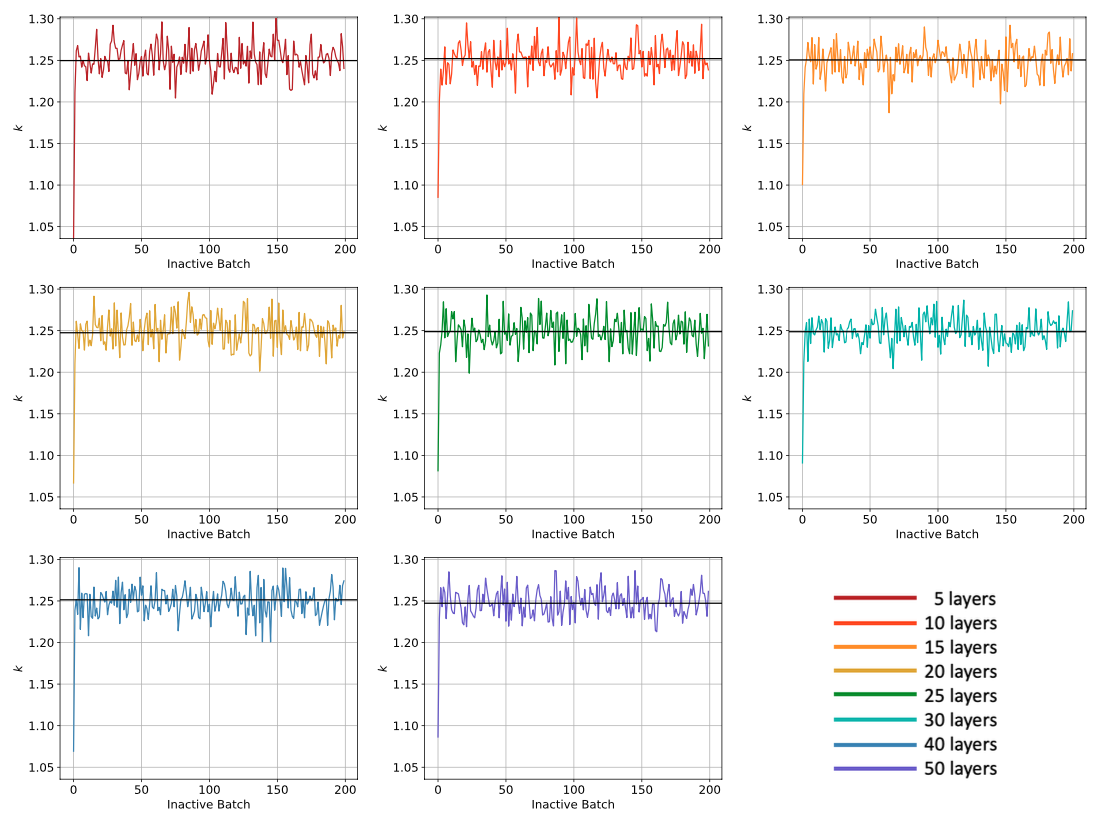
When the script finishes running, plots of Shannon entropy and k, along with the saved statepoint files from the simulations, will be available in a directory named inactive_study. Below are shown example images of the Shannon entropy and k plots. If a method to detect convergence of the fission source is supplied (the default is none), then the script will report whether the chosen metric succeeded or not. Note, there could be various factors affecting whether the metric succeeded or not, and based on user choices, it is possible for a metric to succeed when more batches or more particles per batch are truly needed. These metrics should be considered along side inspection of the plots of the Shannon Entropy, which typically takes longer than the eigenvalue to converge. Having a converged fission source is important so that when the active batches begin tallying, there is no contamination from the source's initial guess impacting the tallies.
Figure 1: Example plots that are generated as part of the inactive study, showing Shannon entropy as a function of inactive batch and number of cell divisions
Figure 2: Example plots that are generated as part of the inactive study, showing k as a function of inactive batch and number of cell divisions; the horizontal line shows the average of k over the last 100 cycles
Cell Discretization
To determine an appropriate cell discretization, you can use the scripts/layers_study.py script. To run this script, you need:
A Python script that creates your OpenMC for you; this Python script must take -n <layers> to indicate the number of cell divisions to create
A Cardinal input file that runs your OpenMC model given some imposed temperature and density distribution
In addition, your Cardinal input file must contain a number of postprocessors, user objects, and vector postprocessors to process the solution to extract the various scalar and averaged quantities used in the convergence study. In the [Problem] block, you must set outputs = 'unrelaxed_tally_std_dev'. You will also need to copy and paste the following into your input file:
warningwarning
When copying the above into your input file, be sure to update the block for the various power postprocessors and user objects to be the blocks on which the heat source is defined, as well as the direction for the LayeredAverage.
For example, to run a cell discretization study for the model developed for the gas compact tutorial, we would run:
since the Python script used to generate the OpenMC model is named unit_cell.py and our Cardinal input file is named openmc.i. You should edit the beginning of the layers_study.py script to select the number of cell layers, number of inactive cycles, and maxium number of active cycles:
import math
use_saved_statepoints = False
n_layers = [10, 20, 30, 40, 50]
n_inactive = 25
n_active = 5
n_max_active = 1000
batch_interval = 2
h = 1.6
average_power_density = 9.605e7
from argparse import ArgumentParser
import multiprocessing
import matplotlib
import matplotlib.pyplot as plt
import pandas
import numpy as np
import openmc
import csv
import sys
import os
matplotlib.rcParams.update({'font.size': 14})
colors = ['firebrick', 'orangered', 'darkorange', 'goldenrod', 'forestgreen', \
'lightseagreen', 'steelblue', 'slateblue']
program_description = ("Script for determining required number of cell divisions "
"for an OpenMC model")
ap = ArgumentParser(description=program_description)
ap.add_argument('-i', dest='script_name', type=str,
help='Name of the OpenMC python script to run (without .py extension)')
ap.add_argument('-n-threads', dest='n_threads', type=int,
default=multiprocessing.cpu_count(), help='Number of threads to run Cardinal with')
ap.add_argument('-input', dest='input_file', type=str,
help='Name of the Cardinal input file to run')
args = ap.parse_args()
input_file = args.input_file
script_name = args.script_name + ".py"
file_path = os.path.realpath(__file__)
file_dir, file_name = os.path.split(file_path)
exec_dir, file_name = os.path.split(file_dir)
methods = ['opt', 'devel', 'oprof', 'dbg']
exec_name = ''
for i in methods:
if (os.path.exists(exec_dir + "/cardinal-" + i)):
exec_name = exec_dir + "/cardinal-" + i
break
if (exec_name == ''):
raise ValueError("No Cardinal executable was found!")
if (not os.path.exists(os.getcwd() + '/layers_study')):
os.makedirs(os.getcwd() + '/layers_study')
nl = len(n_layers)
k = np.empty([nl])
k_std_dev = np.empty([nl])
max_power = np.empty([nl])
min_power = np.empty([nl])
max_power_std_dev = np.empty([nl])
min_power_std_dev = np.empty([nl])
q = np.empty([nl, max(n_layers)])
for i in range(nl):
n = n_layers[i]
file_base = "layers_study/openmc_" + str(n) + "_out"
output_csv = file_base + ".csv"
if ((not use_saved_statepoints) or (not os.path.exists(output_csv))):
os.system("python " + script_name + " -n " + str(n))
os.system(exec_name + " -i " + input_file + \
" UserObjects/average_power_axial/num_layers=" + str(n) + \
" Problem/inactive_batches=" + str(n_inactive) + \
" Problem/batches=" + str(n_active + n_inactive) + \
" Problem/max_batches=" + str(n_max_active + n_inactive) + \
" Problem/batch_interval=" + str(batch_interval) + \
" MultiApps/active='' Transfers/active='' Executioner/num_steps=1" + \
" Outputs/file_base=" + file_base + \
" --n-threads=" + str(args.n_threads))
data = pandas.read_csv(output_csv)
k[i] = data.at[1, 'k']
k_std_dev[i] = data.at[1, 'k_std_dev']
max_power[i] = data.at[1, 'max_power']
min_power[i] = data.at[1, 'min_power']
max_power_std_dev[i] = data.at[1, 'proxy_max_power_std_dev']
min_power_std_dev[i] = data.at[1, 'proxy_min_power_std_dev']
for i in range(1, nl):
n = n_layers[i]
print('Layers: ' + str(n) + ':')
print('\tPercent change in max power: ' + str(abs(max_power[i] - max_power[i - 1]) / max_power[i - 1] * 100))
print('\tPercent change in min power: ' + str(abs(min_power[i] - min_power[i - 1]) / min_power[i - 1] * 100))
print('\tPercent change in max power (rel. to avg.): ' + str(abs(max_power[i] - max_power[i - 1]) / average_power_density * 100))
print('\tPercent change in min power (rel. to avg.): ' + str(abs(min_power[i] - min_power[i - 1]) / average_power_density * 100))
fig, ax = plt.subplots()
plt.errorbar(n_layers, max_power, yerr=max_power_std_dev, fmt='.r', capsize=3.0, marker='o', linestyle='-', color='r')
ax.set_xlabel('Axial Layers')
ax.set_ylabel('Maximum Power (W/m$^3$)')
plt.grid()
plt.savefig('layers_study/' + args.script_name + '_max_power_layers.pdf', bbox_inches='tight')
plt.close()
fig, ax = plt.subplots()
plt.errorbar(n_layers, min_power, yerr=min_power_std_dev, fmt='.b', capsize=3.0, marker='o', linestyle='-', color='b')
ax.set_xlabel('Axial Layers')
ax.set_ylabel('Minimum Power (W/m$^3$)')
plt.grid()
plt.savefig('layers_study/' + args.script_name + '_min_power_layers.pdf', bbox_inches='tight')
plt.close()
fig, ax = plt.subplots()
plt.errorbar(n_layers, k, yerr=k_std_dev, fmt='.k', capsize=3.0, marker='o', color='k', linestyle = '-')
plt.xlabel('Axial Layers')
plt.ylabel('$k$')
plt.grid()
plt.savefig('layers_study/' + args.script_name + '_k_layers.pdf', bbox_inches='tight')
plt.close()
fig, ax = plt.subplots()
for l in range(nl):
i = n_layers[l]
power_file = "layers_study/openmc_" + str(i) + "_out_power_avg_0001.csv"
q = []
with open(power_file) as f:
reader = csv.reader(f)
next(reader)
for row in reader:
q.append(float(row[0]))
dz = h / i
z = np.linspace(dz / 2.0, h - dz / 2.0, i)
plt.plot(z, q, label='{} layers'.format(i), color=colors[l], marker='o', markersize=2.0)
plt.xlabel('Distance from Inlet (m)')
plt.ylabel('Power (W/m$^3$)')
plt.grid()
plt.legend(loc='lower center', ncol=2)
plt.savefig('layers_study/' + args.script_name + '_layers_q.pdf', bbox_inches='tight')
plt.close()
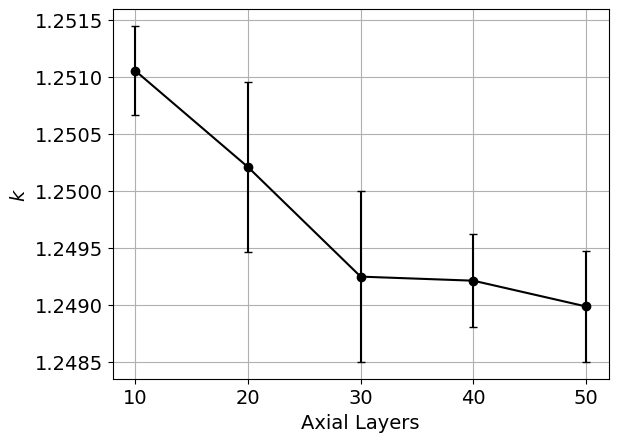
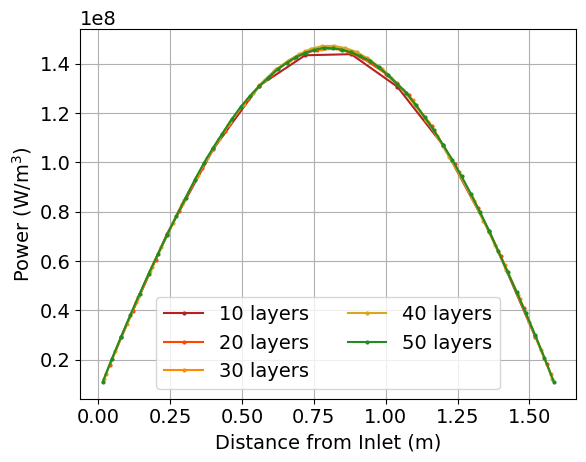
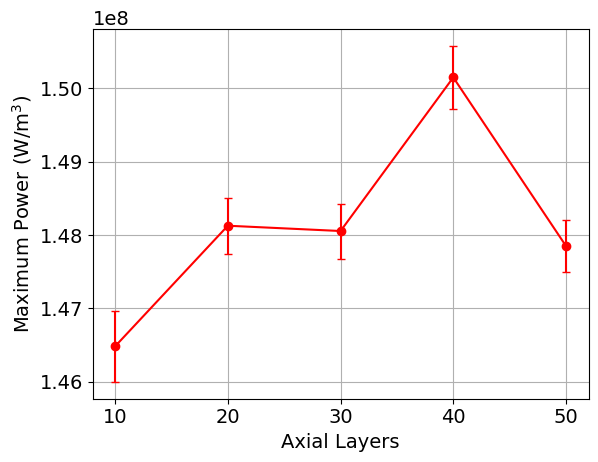
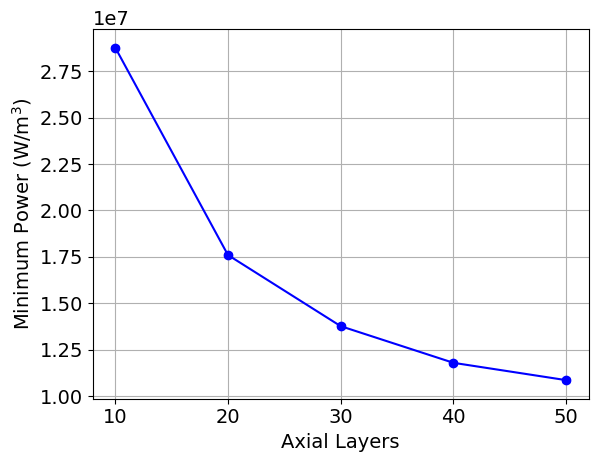
When the script finishes running, plots of the maximum power, minimum power, k, and the axial power distribution, along with the saved CSV output from the simulations, will be available in a directory named layers_study. Below are shown example images of these quantities as a function of the cell discretization. You would then select the cell discretization scheme to be the point at which the changes in power and k are sufficiently converged.
Figure 3: Example plot that is generated as part of the cell discretization study, showing k as a function of the cell discretization
Figure 4: Example plot that is generated as part of the cell discretization study, showing the radially-averaged power distribution as a function of the cell discretization
Figure 5: Example plot that is generated as part of the cell discretization study, showing the maximum power as a function of the cell discretization
Figure 6: Example plot that is generated as part of the cell discretization study, showing the minimum power as a function of the cell discretization