Bayesian Uncertainty Quantification (UQ) on a 1D Diffusion Problem
This example demonstrates how to infer unknown model parameters given experimental observations. Uncertainties of the model parameters are inversely quantified through the Bayesian framework, which makes use of samplers including IndependentGaussianMH (Independent Gaussian Metropolis-Hastings), AffineInvariantStretchSampler or AffineInvariantDES (AffineInvariantDifferential).
Overview of Bayesian UQ
In general, assume a computational model , where are unknown model parameters. Original belief about the unknown parameters, described by the prior distribution , is usually subject to large uncertainties. Given experimental measurements of the computational model at specific configuration conditions , updated knowledge about the model parameters can be obtained by the Bayes' theorem:
where is the Likelihood function. Gaussian and TruncatedGaussian distributions are the most popular choices for the likelihood function. Variance of the Guassian or truncated Gaussian, which describes the sum of the model discrepancy and measurement error (referred to as term herein), can be either fixed a priori or inferred from the experimental measurements . The goal of Bayesian UQ is to infer the unknown parameters (and if needed) given experimental measurements , i.e., the posterior distributions .
The code architecture for performing Markov Chain Monte Carlo (MCMC) sampling in a parallelized fashion is presented in Figure 1. There are three steps for performing the sampling: proposal, model evaluation, and decision making. These three steps and the respective code objects are discussed below:
Proposals:The proposals are made using the user-defined specific MCMC Samplers object which derives from the MCMC base class PMCMCBase. The specific MCMC sampler can be any variant of either serial or parallel MCMC techniques. In total, the MCMC sampler object proposes proposals which are subsequently sent to the MultiApps system.Model evaluation:The MultiApps system executes the models for each of the parallel proposals. In total, there will be model evaluations, where is the number of experimental configurations. MultiApps is responsible for distributing these jobs across a given set of processors.Decision making:The model outputs from the MultiApps system are gathered by a Reporters object to compute the likelihood function. Then, the acceptance probabilities are computed for the parallel proposals and accept/reject decisions are made. The information of accepted samples is sent to the MCMC sampler object to influence the next set of proposals.
The above three steps are repeated for a user-specified number of times.
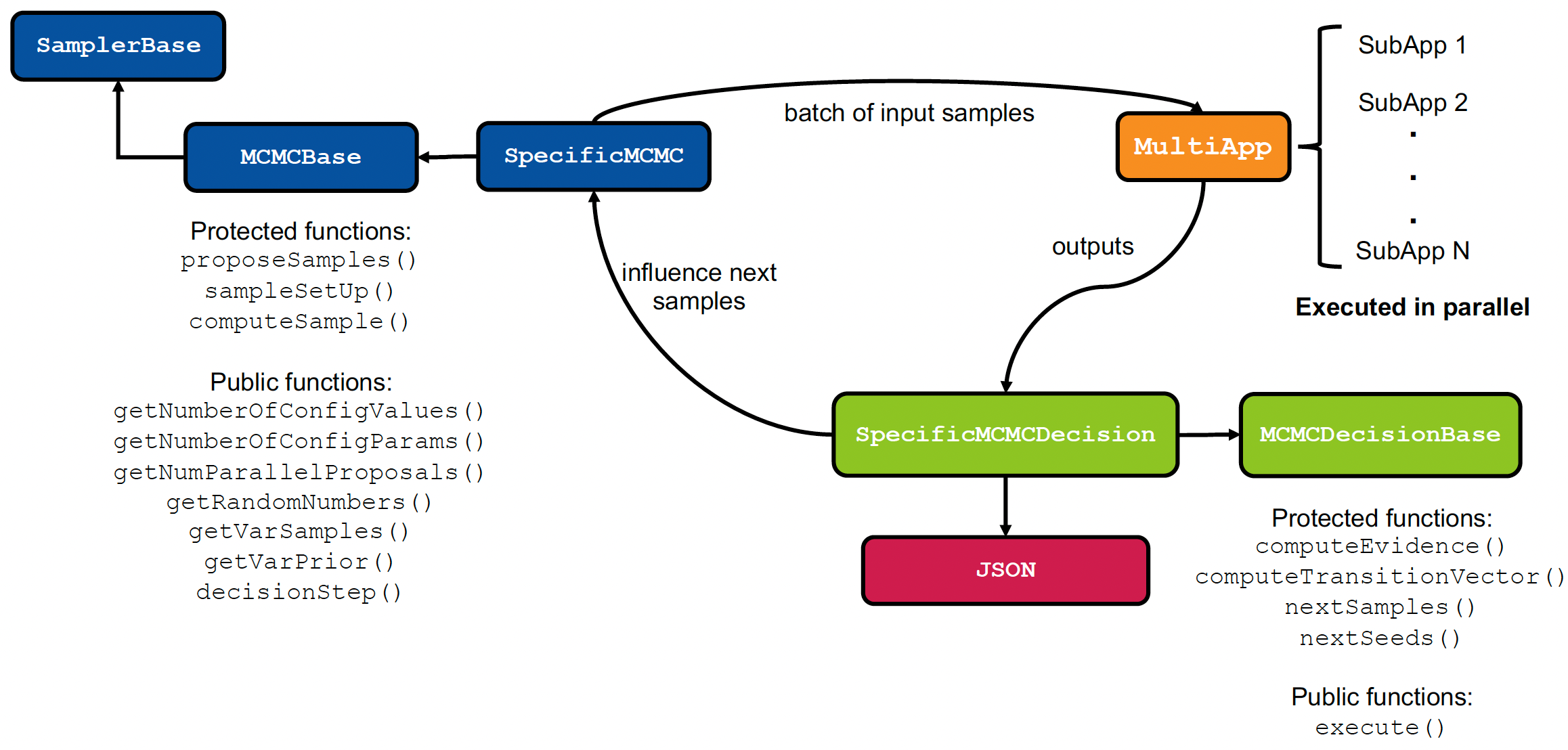
Figure 1: Code architecture for parallelized MCMC sampling in MOOSE for performing Bayesian UQ (Dhulipala et al. (2023)).
Problem Statement
This tutorial demonstrates the application of Bayesian UQ on a 1D transient diffusion problem with Dirichlet boundary conditions on each end of the domain:
where defines the solution field. The experiments measure the average solution field across the entire domain length at the end of time . Experimental measurements are pre-generated at eight configuration points . A Gaussian noise with standard deviation of 0.05 is added to simulate the measurement error. The goal is to infer the lefthandside and righthandside Dirichlet boundary conditions, i.e., and , referred to as and for conciseness. It is worth pointing out that the term, which is the variance of the sum of the experimental measurement and model deviations, can also be inferred during the Bayesian UQ process. This tutorial will first demonstrate the inference of true boundary values by assuming a fixed variance term for the experimental error, followed by the inference of the boundary values together with variance term.
Inferring Calibration Parameters Only (Fixed Term)
Sub-Application Input
The problem defined above, with respect to the MultiApps system, is a sub-application. The complete input file for the problem is provided in Listing 1.
Listing 1: Complete input file for executing the transient diffusion problem.
(contrib/moose/modules/stochastic_tools/test/tests/samplers/mcmc/sub.i)Main Input
The main application, with respect to the MultiApps system, is the driver of the stochastic simulations. The main input does not perform a solve itself, couples with the sub-application for the full stochastic analysis. Details of each individual input section will is explained in details as follows.
The StochasticTools block is defined at the beginning of the main application to automatically create the necessary objects for running a stochastic analysis.
The Distributions block is used to define the prior distributions for the two stochastic boundary conditions. In this example, the two stochastic boundary conditions are assumed to follow normal distributions
A Likelihoods object is created to specify the type of likelihood function used in the analysis.
A Gaussian-type log-likelihood function is used in the current analysis, as specified by
type = Gaussianandlog_likelihood = true.The
noiseterm defines the experimental noise plus model deviations against experiments. Scale of the Gaussian distribution is fixed to a constant value of 0.05 through thenoise_specifiedterm, as will be explained next in the Reporters block. Note that this term can be inferredThe
file_nameargument takes in a CSV file that contains experimental values. In this example, theexp_0.05.csvfile contains 8 independent experimental measurements, which are the sum of pre-simulated solution and a Gaussian noise with standard deviation of 0.05.
In the Samplers block, AffineInvariantDES is used to sample the left and right boundary conditions according to the parallelized Markov chain Monte Carlo method at certain configuration conditions.
prior_distributionstakes in the pre-defined prior distributions of the parameters to be calibrated.num_parallel_proposalsspecifies the number of proposals to make and corresponding sub-applications executed in parallel.initial_valuesdefines the starting values of the parameters to be calibrated.file_nametakes in a CSV file that contains the configuration values. In this example, theconfg.csvfile contains 8 configuration values .
In the MultiApps block, a SamplerFullSolveMultiApp object is defined to create and run a sub-application for each sample provided by the sampler object. More specifically, a full-solve type sub-application is executed for each row of the Sampler matrix.
Under the Transfers block, SamplerReporterTransfer is utilized to transfer data from Reporters on the sub-application to a StochasticReporter on the main application. In this example, the value of a postprocessor named
average, as defined in the sub-application, is transferred to a StochasticReporter namedconstantin the main application.
The Controls block makes use of MultiAppSamplerControl to pass command line arguments from the main application to the sub-applications. In this example, three arguments are passed from the main application to the sub-applications, i.e.,
left_bc: the Dirichlet boundary condition at the lefthandside of the domain.right_bc: the Dirichlet boundary condition at the righthandside of the domain.mesh1: the domain length, which is controlled by the configuration parameters in the Samplers block.
Then, the Reporters section defines
Reporterobjects for data transfer in the MultiApps system.
The
constantreporter of type StochasticReporter serves as a storage container for stochastic simulation results coming from the sub-application.noise_specifiedmakes use of the ConstantReporter reporter to generate a constant value, which is used for the variance term in the experimental error in this example. The value is specified to be 0.05 as defined inreal_values.mcmc_reporteris of type AffineInvariantDES which makes the decision whether or not to accept a proposal generated by the parallel Markov chain Monte Carlo algorithms. It takes in a likelihood functionlikelihoods, asampler, and the model output from the sub-applicationoutput_value. In this example,output_value = constant/reporter_transfer:average:valuemeans that the model output from the sub-application is obtained by a reporter namedconstant, which interacts with the reporterreporter_transferto transfer the average solution field.
Available Sampling Algorithms
Currently, multiple parallel MCMC samplers have been implemented in stochastic tools module, including IndependentGaussianMH, AffineInvariantStretchSampler, and AffineInvariantDES. The above example is demonstrated on AffineInvariantDES. Switching to a different MCMC sampler only requires minor changes in the Samplers block and the Reporters block in the main input file, while the input syntax for the sub-application remains the same. Three available MCMC samplers are explained below respectively:
IndependentGaussianMH(IMH): Performs Metropolis-Hastings MCMC sampling with independent Gaussian proposals. Under the Samplers block,
std_propspecifies the standard deviations of the independent Gaussian proposals for making the next proposal.seed_inputstakes in a reporter namedmcmc_reporterwhich reports seed inputs values for the next proposals. Themcmc_reporteris defined under the Reporters block of type IndependentMHDecision, which performs decision making for independent Metropolis-Hastings MCMC (Calderhead, 2014).
AffineInvariantStretchSampler(SS): Performs affine-invariant ensemble MCMC with stretch sampler (Goodman and Weare, 2010; Shields et al., 2021). Different from other samplers, it takes in two reporters to access the mean and variance of the previous state of all the walkers (
previous_state). By default, the walkers take a step size of 0.2 in the stretch sampler. Correspondingly, anmcmc_reporterof type AffineInvariantStretchDecision need be defined in the Reporters block for decision making.
AffineInvariantDES(DES): Performs affine-invariant ensemble MCMC with differential sampler (Braak, 2006; Nelson et al., 2013). Similar to the stretch sampler, the evolution sampler requires two reporters to access the mean and variance of the previous state of all the walkers (
previous_state). Correspondingly, anmcmc_reporterof type AffineInvariantDifferentialDecision need be defined in the Reporters block for decision making.
Inferring Both Calibration Parameters and the Variance Term
In addition to inferring the model parameters, the variance term can also be inferred at the same time. Both the differential evolution sampler AffineInvariantDES or stretch sampler AffineInvariantStretchSampler can be used for this purpose. Changes in the following blocks are necessary in the main input file.
A prior distribution need be defined under the Distributions block. In this example, a uniform distribution with wide uncertainty range is specified for the variance term as a conservative assumption.
In the definition of the Likelihood block, the variance term for the Gaussian likelihood function is returned by a reporter named
mcmc_reporter, which is defined in the Reporters block of type AffineInvariantDifferentialDecision.
Finally, in the Samplers block, the AffineInvariantDES sampler takes in the prior distribution of the variance term via the
prior_varianceargument. In this example, the prior distribution of the variance term has been defined invarianceunder the Distributions block.
Result and Analysis
The output JSON file contains samples across different processors at different timesteps. Practically, all MCMC samples are generated with serial auto-correlations. Therefore, it is essential to diagnose the sample auto-correlation for predictable UQ quality. Two primary diagnostics are presented here for analyzing the sample quality: the integrated auto-correlation time (), and the effective sample size (ESS) Dhulipala et al. (2023). describes the interval after which a sample can be considered independent given a current sample index, and ESS describes the number of "effective" samples after taking into consideration the sample auto-correlation. To obtain "independent" samples from the Markov chains, the first 100 samples are discarded for "burn-in", with the remaining samples thinned by . The ESS of the generated samples are calculated based on the thinned samples. As presented in Table 1, for IMH, SS and DES decreases, and ESS for the three algorithms increases correspondingly, indicating the best sampling qualify by DES, followed by SS, then IMH.
Table 1: Integrated autocorrelation time () and effective sample size (ESS) when inferring model parameters
| Algorithm | ESS | |
|---|---|---|
| IMH | 123.908 | 9.528 |
| SS | 41.779 | 46.719 |
| DES | 11.217 | 676.547 |
When inferring only the boundary conditions, all three samplers are applied, with 1,000 serial steps simulated with 5 parallel proposals per step. Figure 2 shows the posterior distributions of the calibration parameters and . A strong linearity is observed for the two parameters as would be expected from the underlying physics. The IMH samples appear to be extremely localized compared to DES. The SS samples are almost as good as samples generated by DES in terms of exploring the parameter space. The DES samples are the best from this aspect. More detailed posterior distributions are presented in Figure 3 for all three algorithms IMH, SS and DES. It is generally observed that the posteriors using IMH are restricted in the parameter space. Whereas, the posteriors for both SS and DES are spread across the parameter space and look comparable to each other. This can be observed by comparing Figure 3b and Figure 3c for the posterior of , and Figure 3e and Figure 3f for the posterior of .
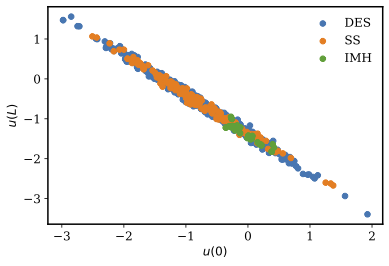
Figure 2: Scatter plot of the samples from IMH, SS, and DES samplers when only inferring the model parameters.
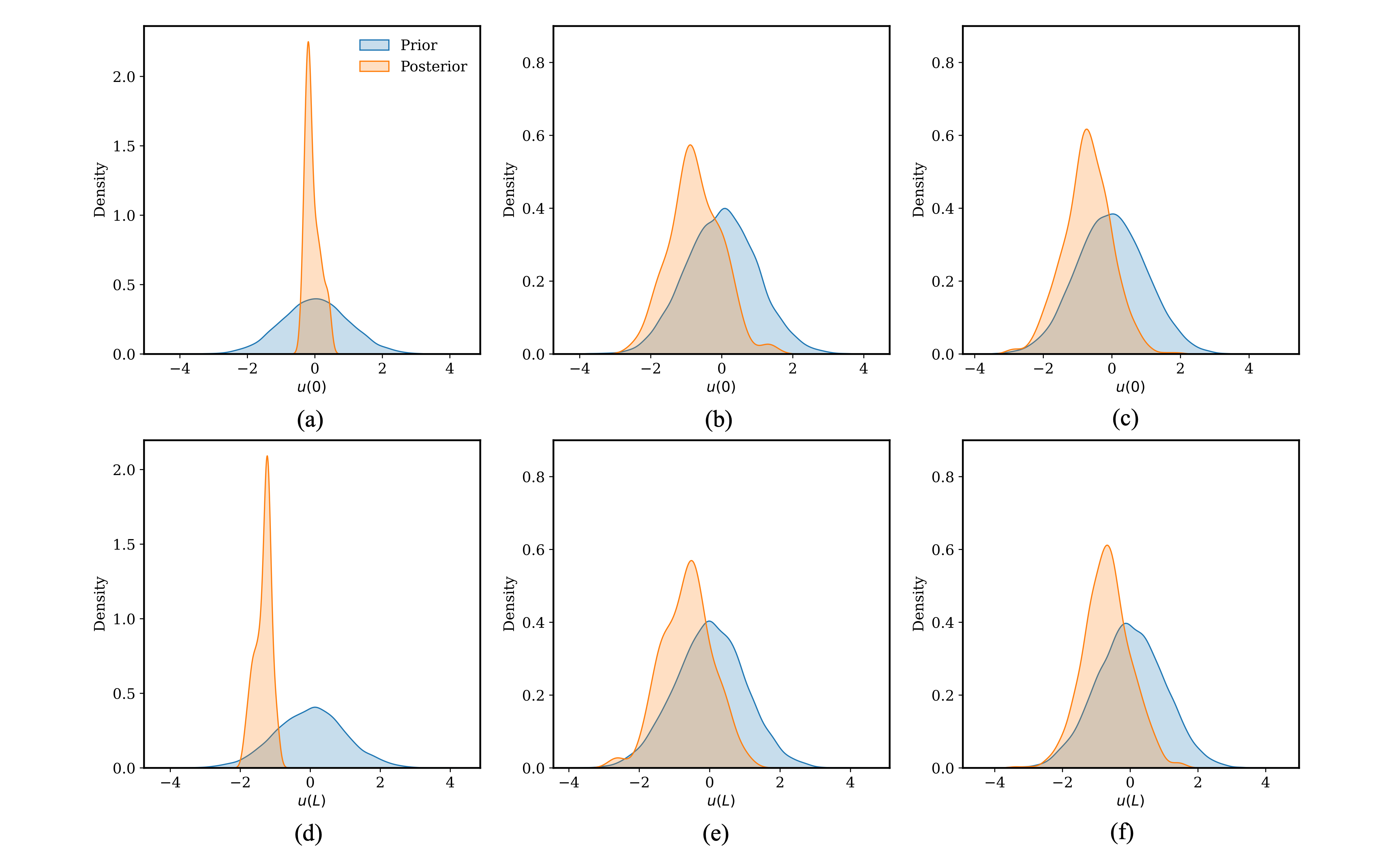
Figure 3: Posterior distributions for the diffusion time derivative problem when inferring only the model parameters. (a), (b) and (c) are the posteriors using IMH, SS, and DES samplers, respectively. (d), (e) and (f) are the posteriors using IMH, SS, and DES samplers, respectively.
When inferring both boundary conditions and the term, only the SS and DES samplers are considered given the poor performance of the IMH sampler evidenced from the above case. Again, 1,000 serial steps are simulated with 5 parallel proposals per step. Sampling diagnostics are presented for the two samplers in Table 2. The for both samplers are comparable, and the for DES increased compared to the previous case of inferring only model parameters, possible caused by the increased problem complexity due to the inclusion of the term. For the ESS, the DES shows a three time enhancement compared to SS, due to the higher residual sample autocorrelation upon chain thinning.
Table 2: Integrated autocorrelation time () and effective sample size (ESS) when inferring both model parameters and the term.
| Algorithm | ESS | |
|---|---|---|
| SS | 36.228 | 51.991 |
| DES | 45.542 | 155.0 |
Scatter plot of the inferred model parameters and is presented in Figure 4. Again, the two model parameters exhibit almost a perfect correlation which is consistent with the observation in the previous case, due to the ill-posedness of the problem.
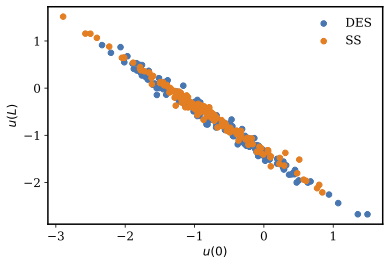
Figure 4: Scatter plot of the samples from IMH, SS, and DES samplers when inferring both model parameters and the term.
Detailed posterior distributions of , and the term are presented in Figure 5 for the SS and DES samplers. Both samplers are producing quite consistent posterior distributions. Specifically, Figure 5a and Figure 5d need to be compared for , Figure 5b and Figure 5e for , and Figure 5c and Figure 5f for the term. Both samplers produce consistent posterior distributions, and the posterior distributions of the term are approximately centered around the true value of the Gaussian noise (denoted by the solid vertical line) that was added to replicate the “experimental” data.
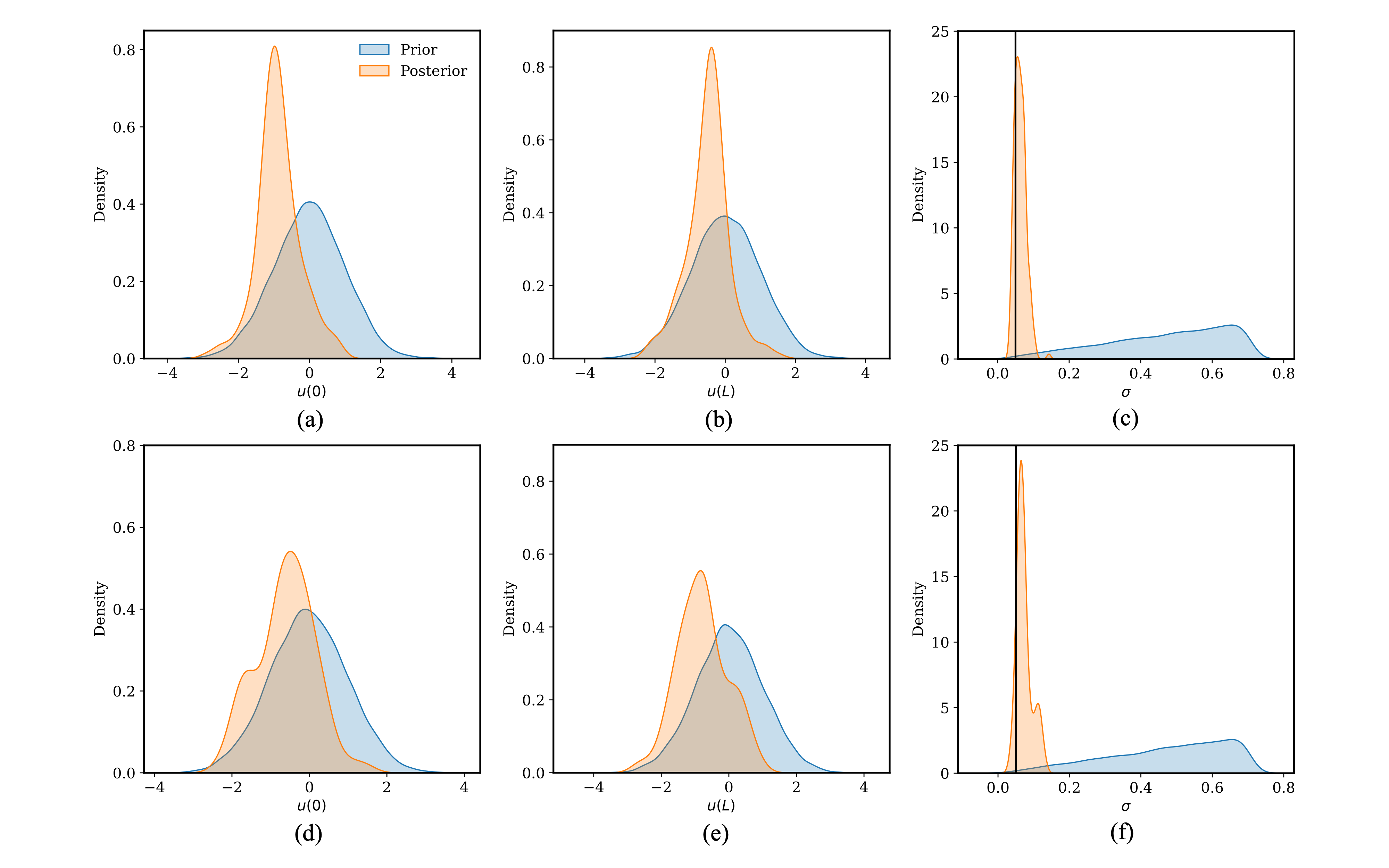
Figure 5: Posterior distributions for the diffusion time derivative problem when inferring the model parameters and the term. (a), (b) and (c) show the posteriors using SS. (d), (e) and (f) show the posteriors using DES.
As shown in Figure 3 and Figure 5, posteriors of the model parameters and potentially the term are much more constrained compared to the priors, reflecting the integration of experimental information into the models through the Bayesian UQ process. Followup forward UQ based on the updated posteriors will provide more accurate information about the output uncertainty.
References
- C. J. T. Braak.
A markov chain monte carlo version of the genetic algorithm differential evolution: easy bayesian computing for real parameter spaces.
Statistics and Computing, 16:239–249, 2006.
doi:10.1007/s11222-006-8769-1.[BibTeX]
- B. Calderhead.
A general construction for parallelizing metropolis-hastings algorithms.
Proceedings of the National Academy of Sciences, 111(49):17408–17413, 2014.
doi:10.1073/pnas.1408184111.[BibTeX]
- Som LakshmiNarasimha Dhulipala, Daniel Schwen, Yifeng Che, Ryan Terrence Sweet, Aysenur Toptan, Zachary M Prince, Peter German, and Stephen R Novascone.
Massively parallel bayesian model calibration and uncertainty quantification with applications to nuclear fuels and materials.
Technical Report, Idaho National Laboratory (INL), Idaho Falls, ID (United States), 2023.[BibTeX]
- J. Goodman and J. Weare.
Ensemble samplers with affine invariance.
Communications in applied mathematics and computational science, 5(1):65–80, 2010.
doi:10.2140/camcos.2010.5.65.[BibTeX]
- B. Nelson, E. B. Ford, and M. J. Payne.
Run dmc: an efficient, parallel code for analyzing radial velocity observations using n-body integrations and differential evolution markov chain monte carlo.
The Astrophysical Journal Supplement Series, 11:11–25, 2013.
doi:10.1088/0067-0049/210/1/11.[BibTeX]
- M. D. Shields, D. G. Giovanis, and V. S. Sundar.
Subset simulation for problems with strongly non-gaussian, highly anisotropic, and degenerate distributions.
Computers & Structures, 245:106431, 2021.
doi:10.1016/j.compstruc.2020.106431.[BibTeX]